MacroAPE 1083:ETV1 GIST48-ETV1-ChIP-Seq/Homer
From FANTOM5_SSTAR
Full Name: ETV1_GIST48-ETV1-ChIP-Seq/Homer
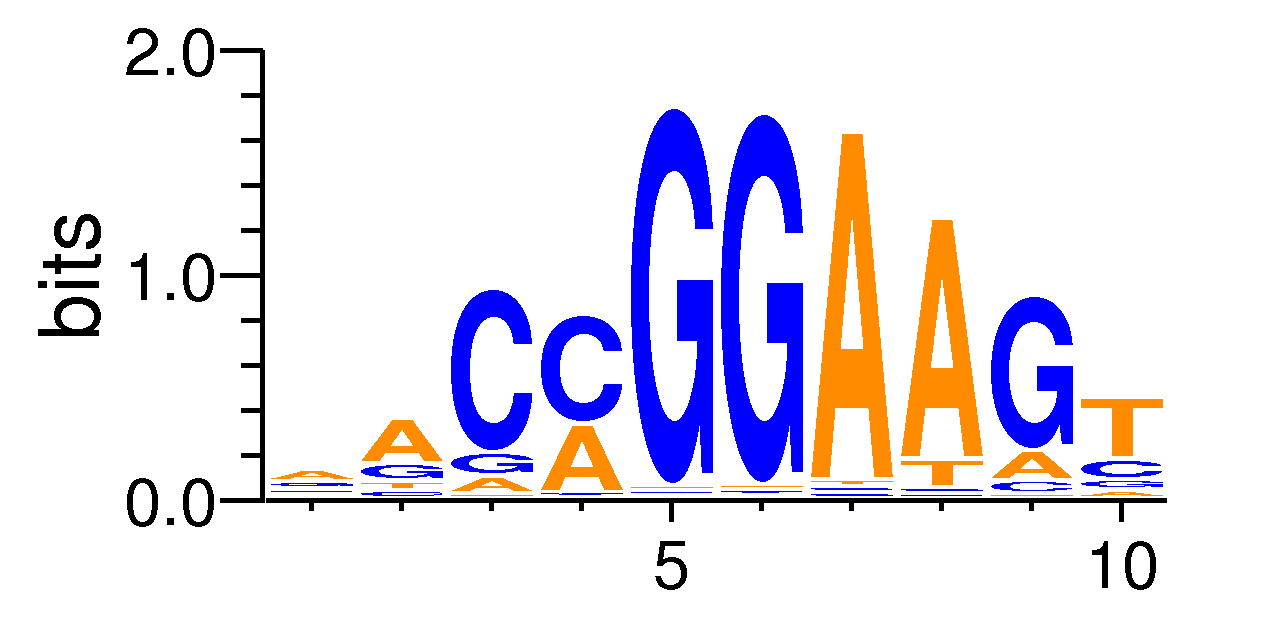
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| ELK4#MA0076.1 | -1 | 3.43735e-08 | 1.63618e-05 | 3.21625e-05 | 9 | AACCGGAAGT | ACCGGAAGT | + |
| Eip74EF#MA0026.1 | -2 | 9.04007e-08 | 4.30307e-05 | 4.2293e-05 | 7 | AACCGGAAGT | CCGGAAG | + |
| GABPA#MA0062.2 | -1 | 3.90676e-06 | 0.00185962 | 0.00121849 | 9 | AACCGGAAGT | ACCGGAAGAG | + |
| ELK1#MA0028.1 | 1 | 1.34104e-05 | 0.00638334 | 0.00313695 | 9 | AACCGGAAGT | GAGCCGGAAG | + |
| FEV#MA0156.1 | -2 | 1.78566e-05 | 0.00849976 | 0.00334161 | 8 | AACCGGAAGT | CAGGAAAT | + |
| GABPA#MA0062.2 | -2 | 2.8279e-05 | 0.0134608 | 0.00441001 | 8 | AACCGGAAGT | CCGGAAGTGGC | + |
| SPI1#MA0080.2 | -3 | 0.000154631 | 0.0736042 | 0.0206692 | 7 | AACCGGAAGT | AGGAAGT | + |
| ELF5#MA0136.1 | -2 | 0.000278729 | 0.132675 | 0.0326001 | 8 | AACCGGAAGT | AAGGAAGTA | - |
| SPI1#MA0080.2 | -3 | 0.000328229 | 0.156237 | 0.0327442 | 6 | AACCGGAAGT | CGGAAG | + |
| IXR1#MA0323.1 | 1 | 0.000349952 | 0.166577 | 0.0327442 | 10 | AACCGGAAGT | AAGCCGGAAGCGGGG | + |
| ETS1#MA0098.1 | -3 | 0.000505273 | 0.24051 | 0.0429794 | 6 | AACCGGAAGT | CGGAAA | - |
| LYS14#MA0325.1 | -2 | 0.000637491 | 0.303446 | 0.0462215 | 8 | AACCGGAAGT | CCGGAATT | + |
| ASG1#MA0275.1 | -2 | 0.000642185 | 0.30568 | 0.0462215 | 6 | AACCGGAAGT | CCGGAA | + |
| SPIB#MA0081.1 | -1 | 0.000771238 | 0.367109 | 0.051545 | 7 | AACCGGAAGT | AGAGGAA | + |
| YBR239C#MA0420.1 | -1 | 0.00093645 | 0.44575 | 0.0584144 | 8 | AACCGGAAGT | ACCGGAAC | + |
| CAT8#MA0280.1 | -2 | 0.00119256 | 0.567658 | 0.0697407 | 6 | AACCGGAAGT | CCGGAA | + |
| YKL222C#MA0428.1 | -1 | 0.00279944 | 1.33254 | 0.154081 | 9 | AACCGGAAGT | AACGGAAAT | + |
| YRM1#MA0438.1 | -2 | 0.00391563 | 1.86384 | 0.188499 | 8 | AACCGGAAGT | ACGGAAAT | + |
| GCR2#MA0305.1 | -3 | 0.00402146 | 1.91422 | 0.188499 | 7 | AACCGGAAGT | AGGAAGC | - |
| HAL9#MA0311.1 | -3 | 0.00402913 | 1.91787 | 0.188499 | 5 | AACCGGAAGT | CGGAA | + |
| YJL103C#MA0427.1 | 1 | 0.00451222 | 2.14781 | 0.201047 | 10 | AACCGGAAGT | TCTCCGGAGCT | - |
| YER184C#MA0424.1 | 0 | 0.00538477 | 2.56315 | 0.229019 | 8 | AACCGGAAGT | CTCCGGAA | + |
| Stat3#MA0144.1 | 1 | 0.00846996 | 4.0317 | 0.344572 | 9 | AACCGGAAGT | TTCCAGGAAG | + |
| PEND#MA0127.1 | 0 | 0.0107226 | 5.10396 | 0.418038 | 10 | AACCGGAAGT | AATAAGAAGT | - |
| ECM22#MA0292.1 | 0 | 0.0113884 | 5.42088 | 0.426236 | 7 | AACCGGAAGT | CTCCGGA | + |
| STAT1#MA0137.2 | 4 | 0.0152619 | 7.26465 | 0.515044 | 10 | AACCGGAAGT | CATTTCCCGGAAACC | + |
| SIP4#MA0380.1 | -1 | 0.0154126 | 7.33639 | 0.515044 | 7 | AACCGGAAGT | TCCGGAG | - |
| RDR1#MA0360.1 | -1 | 0.018206 | 8.66607 | 0.567833 | 8 | AACCGGAAGT | TGCGGAAC | + |