MacroAPE 1083:5-TCTGCGTCAC
From FANTOM5_SSTAR
Full Name: 5-TCTGCGTCAC,CNhs13223,BestGuess:CREB1.p2.SwissRegulon.nucfreq(0.738713041987632),T:571.0(22.94%),B:5615.5(12.67%),P:1e-44
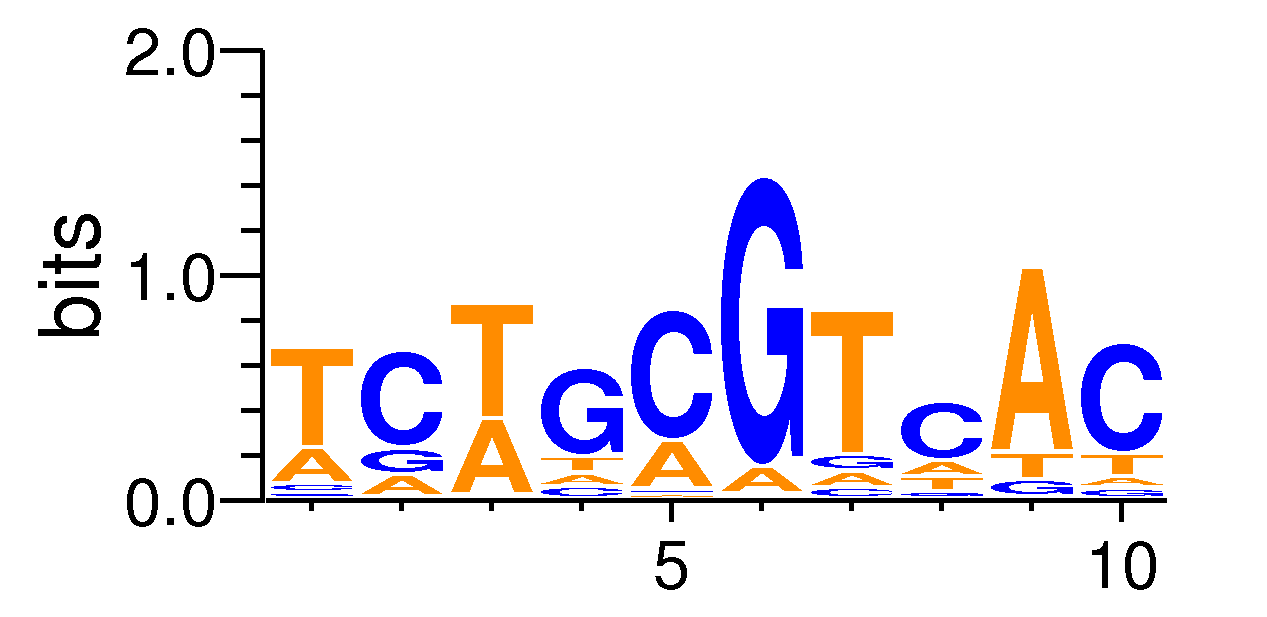
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| FHL1#MA0295.1 | 0 | 0.000654241 | 0.311419 | 0.29033 | 8 | TCTGCGTCAC | TTTGCGTC | - |
| ARG81#MA0272.1 | -2 | 0.000753072 | 0.358462 | 0.29033 | 8 | TCTGCGTCAC | AGAGTCAC | - |
| ARG80#MA0271.1 | -3 | 0.000919274 | 0.437575 | 0.29033 | 6 | TCTGCGTCAC | GCGTCT | - |
| NFE2L2#MA0150.1 | 1 | 0.00251481 | 1.19705 | 0.562266 | 10 | TCTGCGTCAC | TGCTGAGTCAT | - |
| Fos#MA0099.1 | -2 | 0.0032893 | 1.56571 | 0.562266 | 8 | TCTGCGTCAC | TGACTCAC | - |
| AP1#MA0099.2 | -2 | 0.00361202 | 1.71932 | 0.562266 | 7 | TCTGCGTCAC | TGAGTCA | - |
| Sox2#MA0143.1 | 0 | 0.00450763 | 2.14563 | 0.562266 | 10 | TCTGCGTCAC | TTTGCATAACAAAGG | - |
| CST6#MA0286.1 | -1 | 0.00503861 | 2.39838 | 0.562266 | 9 | TCTGCGTCAC | TTACGTCAT | - |
| TGA1A#MA0129.1 | -2 | 0.00586905 | 2.79367 | 0.562266 | 7 | TCTGCGTCAC | TACGTCA | + |
| CRZ1#MA0285.1 | -1 | 0.00593436 | 2.82476 | 0.562266 | 9 | TCTGCGTCAC | CTAAGCCAC | + |
| CREB1#MA0018.1 | -2 | 0.00859725 | 4.09229 | 0.600908 | 8 | TCTGCGTCAC | GACGTCACCAGG | - |
| Pax2#MA0067.1 | -3 | 0.00945825 | 4.50213 | 0.600908 | 7 | TCTGCGTCAC | GCGTGACT | - |
| PPARG#MA0066.1 | 0 | 0.0102182 | 4.86385 | 0.600908 | 10 | TCTGCGTCAC | AGTAGGTCACCGTGACCTAC | - |
| Mafb#MA0117.1 | -3 | 0.0105328 | 5.01363 | 0.600908 | 7 | TCTGCGTCAC | GCGTCAGC | - |
| Pou5f1#MA0142.1 | 1 | 0.0107818 | 5.13211 | 0.600908 | 10 | TCTGCGTCAC | ATTTGCATAACAAAG | - |
| ESR1#MA0112.2 | 3 | 0.012523 | 5.96097 | 0.64679 | 10 | TCTGCGTCAC | AGGTCAGGGTGACCTGGGCC | - |
| CUP9#MA0288.1 | 0 | 0.0129703 | 6.17386 | 0.64679 | 9 | TCTGCGTCAC | AATGTGTCA | - |
| RORA_1#MA0071.1 | 1 | 0.0152519 | 7.25992 | 0.722541 | 9 | TCTGCGTCAC | ATCAAGGTCA | + |
| ESR2#MA0258.1 | -1 | 0.0170277 | 8.10517 | 0.733332 | 9 | TCTGCGTCAC | CAAGGTCACGGTGACCTG | + |
| GCN4#MA0303.1 | 5 | 0.0191757 | 9.12765 | 0.745134 | 10 | TCTGCGTCAC | CAAGGGATGAGTCATACTTCA | + |
| RXR::RAR_DR5#MA0159.1 | 5 | 0.0194166 | 9.24229 | 0.745134 | 10 | TCTGCGTCAC | TGACCTCTCCGTGACCT | - |
| Tal1::Gata1#MA0140.1 | 4 | 0.0205815 | 9.7968 | 0.745134 | 10 | TCTGCGTCAC | CTTATCTGTCCCCACCAG | - |
| bZIP910#MA0096.1 | -3 | 0.0206511 | 9.82991 | 0.745134 | 7 | TCTGCGTCAC | ACGTCAT | - |