MacroAPE 1083:Motif316
From FANTOM5_SSTAR
Full Name: motif316
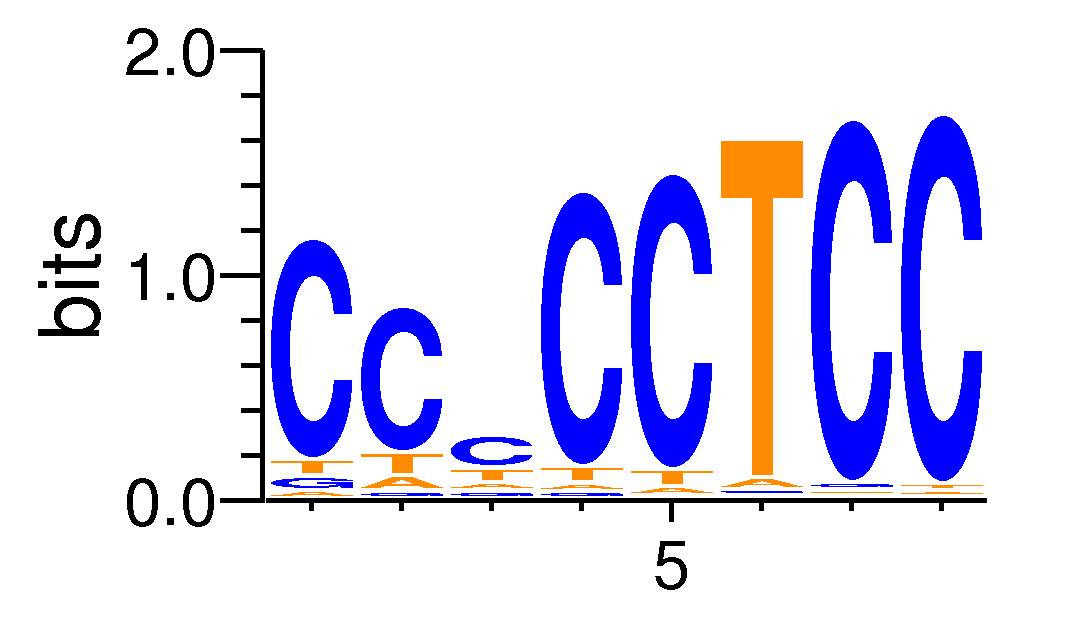
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| MZF1_5-13#MA0057.1 | 2 | 0.000112117 | 0.0533677 | 0.10597 | 8 | CCCCCTCC | TTCCCCCTAC | - |
| SP1#MA0079.2 | 2 | 0.000485994 | 0.231333 | 0.229675 | 8 | CCCCCTCC | CCCCGCCCCC | + |
| RGM1#MA0366.1 | -1 | 0.00245623 | 1.16917 | 0.574184 | 5 | CCCCCTCC | CCCCT | - |
| MSN4#MA0342.1 | -1 | 0.00276403 | 1.31568 | 0.574184 | 5 | CCCCCTCC | CCCCT | - |
| YML081W#MA0431.1 | 0 | 0.00303744 | 1.44582 | 0.574184 | 8 | CCCCCTCC | ACCCCGCAC | + |
| MSN2#MA0341.1 | -1 | 0.00419965 | 1.99903 | 0.632375 | 5 | CCCCCTCC | CCCCT | - |
| YGR067C#MA0425.1 | 0 | 0.00641247 | 3.05233 | 0.632375 | 8 | CCCCCTCC | ACCCCACTTTTTTG | + |
| Pax4#MA0068.1 | 22 | 0.00724391 | 3.4481 | 0.632375 | 8 | CCCCCTCC | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| RREB1#MA0073.1 | 10 | 0.00813321 | 3.87141 | 0.632375 | 8 | CCCCCTCC | CCCCAAACCACCCCCCCCCC | + |
| MIG1#MA0337.1 | 0 | 0.00814875 | 3.87881 | 0.632375 | 7 | CCCCCTCC | CCCCCGC | + |
| GABPA#MA0062.2 | 1 | 0.00853422 | 4.06229 | 0.632375 | 8 | CCCCCTCC | GCCACTTCCGG | - |
| CHA4#MA0283.1 | 0 | 0.00863126 | 4.10848 | 0.632375 | 8 | CCCCCTCC | TCTCCGCC | - |
| YPR022C#MA0436.1 | -1 | 0.009532 | 4.53723 | 0.632375 | 7 | CCCCCTCC | CCCCACG | + |
| opa#MA0456.1 | 2 | 0.0100756 | 4.79598 | 0.632375 | 8 | CCCCCTCC | GACCCCCCGCTG | + |
| ADR1#MA0268.1 | 0 | 0.0107049 | 5.09552 | 0.632375 | 7 | CCCCCTCC | ACCCCAC | + |
| MIG2#MA0338.1 | -1 | 0.0107049 | 5.09552 | 0.632375 | 7 | CCCCCTCC | CCCCGCA | + |
| CTCF#MA0139.1 | 5 | 0.0121165 | 5.76748 | 0.643668 | 8 | CCCCCTCC | TAGCGCCCCCTGGTGGCCA | - |
| btd#MA0443.1 | 1 | 0.0129313 | 6.15531 | 0.643668 | 8 | CCCCCTCC | TCCGCCCCCT | - |
| MIG3#MA0339.1 | -1 | 0.012939 | 6.15898 | 0.643668 | 7 | CCCCCTCC | CCCCGCA | + |
| SP1#MA0079.2 | 1 | 0.0139098 | 6.62108 | 0.64915 | 8 | CCCCCTCC | ACCCCGCCCC | - |
| Klf4#MA0039.2 | 0 | 0.0144228 | 6.86527 | 0.64915 | 8 | CCCCCTCC | GCCCCACCCA | - |
| IXR1#MA0323.1 | 1 | 0.0194275 | 9.24748 | 0.834655 | 8 | CCCCCTCC | CCCCGCTTCCGGCTT | - |
| ZMS1#MA0441.1 | 1 | 0.0203791 | 9.70047 | 0.837474 | 8 | CCCCCTCC | TTCCCCGCA | + |