MacroAPE 1083:CNhs10625 bl f0
From FANTOM5_SSTAR
Full Name: CNhs10625_bl_f0
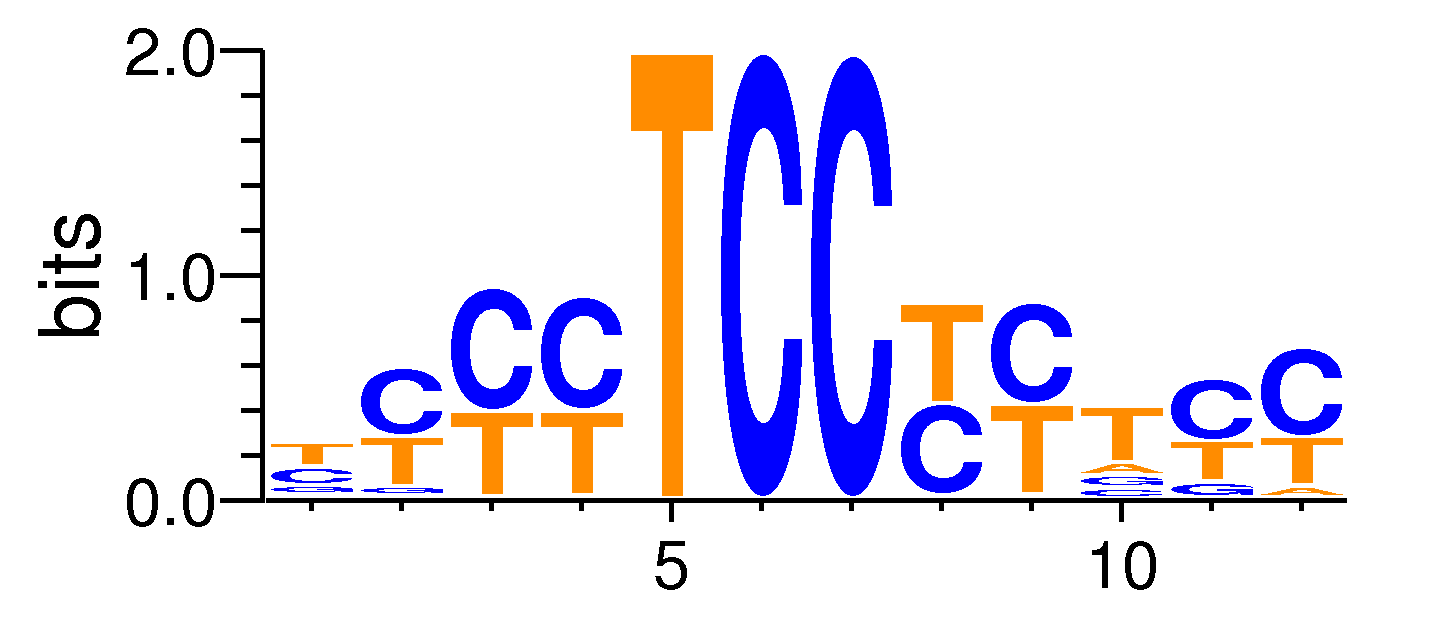
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| Klf4#MA0039.1 | -1 | 9.99678e-05 | 0.0475847 | 0.0951694 | 10 | TCCCTCCTCTCC | CCTTCCTTTA | - |
| id1#MA0120.1 | -1 | 0.00161005 | 0.766382 | 0.628317 | 11 | TCCCTCCTCTCC | TTTTCCTTTTCG | + |
| SPIB#MA0081.1 | -3 | 0.00197999 | 0.942475 | 0.628317 | 7 | TCCCTCCTCTCC | TTCCTCT | - |
| MZF1_5-13#MA0057.1 | -2 | 0.00494469 | 2.35367 | 0.853124 | 10 | TCCCTCCTCTCC | TTCCCCCTAC | - |
| MZF1_1-4#MA0056.1 | -4 | 0.00529365 | 2.51978 | 0.853124 | 6 | TCCCTCCTCTCC | TCCCCA | - |
| HMG-I/Y#MA0045.1 | 1 | 0.00660386 | 3.14344 | 0.853124 | 12 | TCCCTCCTCTCC | GTTTTTCCATTTGTTG | - |
| SP1#MA0079.2 | 0 | 0.00677345 | 3.22416 | 0.853124 | 10 | TCCCTCCTCTCC | CCCCGCCCCC | + |
| SPI1#MA0080.1 | -2 | 0.00764167 | 3.63744 | 0.853124 | 6 | TCCCTCCTCTCC | CTTCCG | - |
| Pax4#MA0068.1 | 17 | 0.00806525 | 3.83906 | 0.853124 | 12 | TCCCTCCTCTCC | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| ZMS1#MA0441.1 | -3 | 0.00971954 | 4.6265 | 0.890809 | 9 | TCCCTCCTCTCC | TTCCCCGCA | + |
| ETS1#MA0098.1 | -2 | 0.0114814 | 5.46514 | 0.890809 | 6 | TCCCTCCTCTCC | CTTCCG | + |
| YGR067C#MA0425.1 | 1 | 0.0115226 | 5.48474 | 0.890809 | 12 | TCCCTCCTCTCC | ACCCCACTTTTTTG | + |
| RME1#MA0370.1 | -3 | 0.0130611 | 6.21709 | 0.890809 | 9 | TCCCTCCTCTCC | TTCCTTTGGA | - |
| ELF5#MA0136.1 | 0 | 0.0131001 | 6.23566 | 0.890809 | 9 | TCCCTCCTCTCC | TACTTCCTT | + |
| EWSR1-FLI1#MA0149.1 | 2 | 0.0143344 | 6.82316 | 0.909754 | 12 | TCCCTCCTCTCC | CCTTCCTTCCTTCCTTCC | - |
| Trl#MA0205.1 | -3 | 0.0154872 | 7.37191 | 0.921489 | 9 | TCCCTCCTCTCC | TTGCTCTCTC | + |
| PPARG::RXRA#MA0065.2 | 3 | 0.0181812 | 8.65427 | 0.971099 | 12 | TCCCTCCTCTCC | TGACCTTTGCCCTAC | - |
| FOXC1#MA0032.1 | -4 | 0.0195704 | 9.31549 | 0.971099 | 8 | TCCCTCCTCTCC | TACTTACC | - |
| ELK1#MA0028.1 | -2 | 0.0204012 | 9.71099 | 0.971099 | 10 | TCCCTCCTCTCC | CTTCCGGCTC | - |
| btd#MA0443.1 | -1 | 0.0204012 | 9.71099 | 0.971099 | 10 | TCCCTCCTCTCC | TCCGCCCCCT | - |