MacroAPE 1083:CNhs12087 bu f1
From FANTOM5_SSTAR
Full Name: CNhs12087_bu_f1
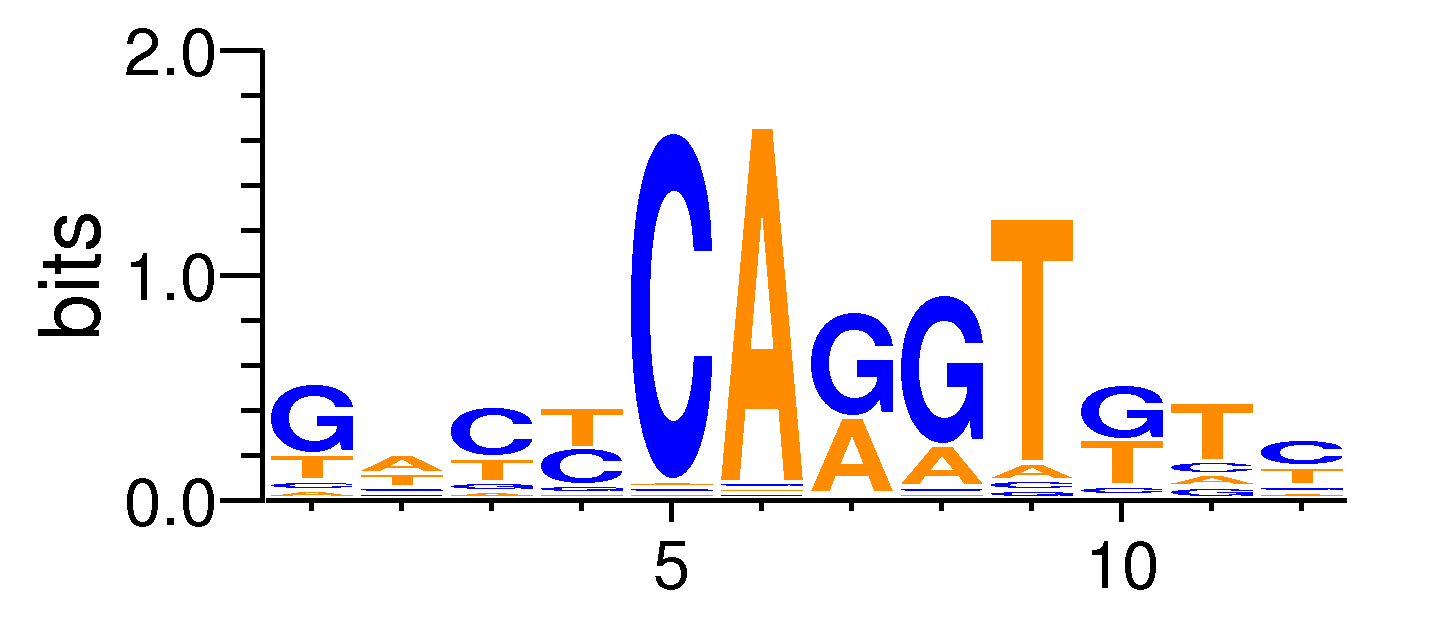
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| sna#MA0086.1 | -4 | 0.000124983 | 0.0594921 | 0.118703 | 6 | GACTCAGGTGTC | CAGGTG | + |
| tin#MA0247.1 | -2 | 0.00109678 | 0.522068 | 0.404352 | 8 | GACTCAGGTGTC | CTCAAGTG | + |
| ARR1#MA0274.1 | -1 | 0.00127724 | 0.607965 | 0.404352 | 8 | GACTCAGGTGTC | ATTCAAGT | - |
| ZEB1#MA0103.1 | -4 | 0.00244533 | 1.16398 | 0.49492 | 6 | GACTCAGGTGTC | CAGGTG | - |
| vnd#MA0253.1 | -1 | 0.00260553 | 1.24023 | 0.49492 | 9 | GACTCAGGTGTC | TTTCAAGTG | + |
| ceh-22#MA0264.1 | -1 | 0.00314238 | 1.49577 | 0.497412 | 11 | GACTCAGGTGTC | TTTCAAGTGGT | - |
| ESR1#MA0112.2 | 0 | 0.00374243 | 1.7814 | 0.507767 | 12 | GACTCAGGTGTC | GGCCCAGGTCACCCTGACCT | + |
| HAC1#MA0310.1 | -4 | 0.00651024 | 3.09888 | 0.772887 | 8 | GACTCAGGTGTC | CACGTGTC | - |
| MAX#MA0058.1 | -3 | 0.0113367 | 5.39625 | 0.997635 | 9 | GACTCAGGTGTC | TCACGTGGTC | - |
| EmBP-1#MA0128.1 | -3 | 0.0119546 | 5.69037 | 0.997635 | 8 | GACTCAGGTGTC | CCACGTGT | - |
| YJL103C#MA0427.1 | -1 | 0.0142496 | 6.78283 | 0.997635 | 11 | GACTCAGGTGTC | TCTCCGGAGCT | - |
| twi#MA0249.1 | -1 | 0.0150767 | 7.1765 | 0.997635 | 11 | GACTCAGGTGTC | TCGCATATGTTG | + |
| Arnt#MA0004.1 | -4 | 0.0161564 | 7.69046 | 0.997635 | 6 | GACTCAGGTGTC | CACGTG | - |
| Mycn#MA0104.2 | -2 | 0.0192389 | 9.15773 | 0.997635 | 10 | GACTCAGGTGTC | CGCACGTGGC | + |
| USF1#MA0093.1 | -3 | 0.020015 | 9.52713 | 0.997635 | 7 | GACTCAGGTGTC | CCACGTG | - |