MacroAPE 1083:PURA f1
From FANTOM5_SSTAR
Full Name: PURA_f1
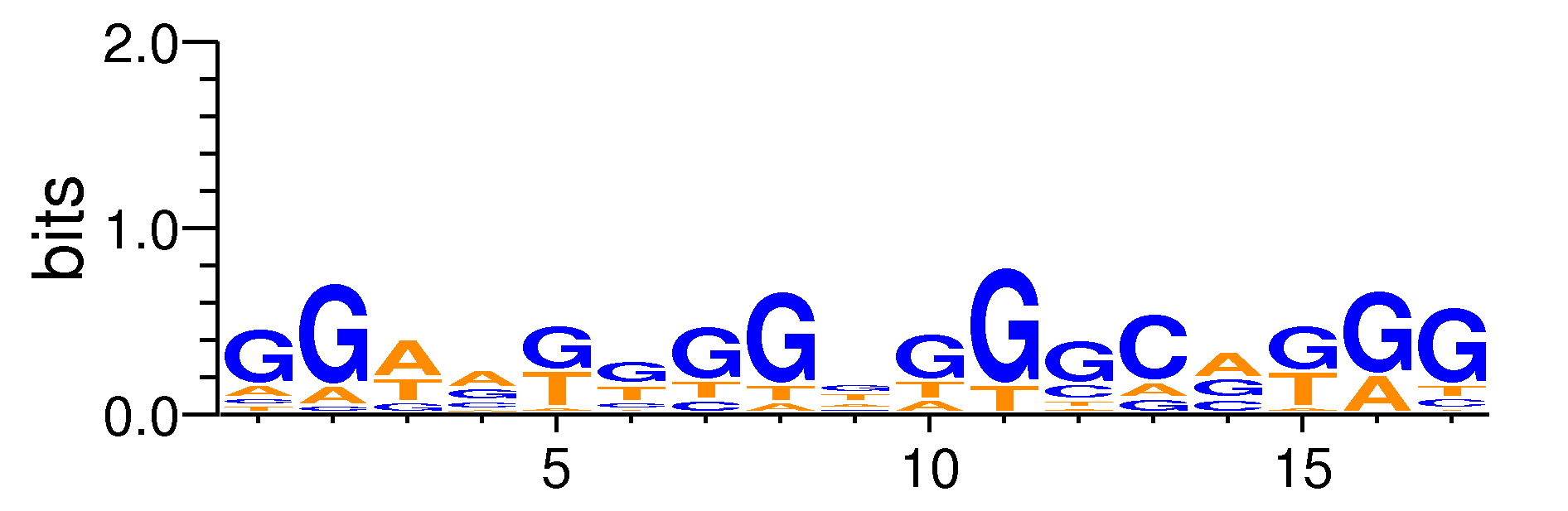
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| SP1#MA0079.2 | -7 | 4.09809e-06 | 0.00195069 | 0.00387568 | 10 | GGAAGGGGGGGGCAGGG | GGGGGCGGGG | - |
| Pax4#MA0068.1 | 1 | 1.60612e-05 | 0.00764514 | 0.00759478 | 17 | GGAAGGGGGGGGCAGGG | GGGGGGGGAGTGGAGTATTGGAAATTTTTC | - |
| SP1#MA0079.2 | -8 | 6.80813e-05 | 0.0324067 | 0.0214622 | 9 | GGAAGGGGGGGGCAGGG | GGGGCGGGGT | + |
| btd#MA0443.1 | -6 | 0.000164852 | 0.0784695 | 0.0389763 | 10 | GGAAGGGGGGGGCAGGG | AGGGGGCGGA | + |
| RREB1#MA0073.1 | 1 | 0.000263395 | 0.125376 | 0.0498201 | 17 | GGAAGGGGGGGGCAGGG | TGGGGGGGGGTGGTTTGGGG | - |
| Macho-1#MA0118.1 | -5 | 0.000447651 | 0.213082 | 0.0705594 | 9 | GGAAGGGGGGGGCAGGG | TGGGGGGTC | + |
| Klf4#MA0039.2 | -8 | 0.00075216 | 0.358028 | 0.10162 | 9 | GGAAGGGGGGGGCAGGG | TGGGTGGGGC | + |
| YGR067C#MA0425.1 | 0 | 0.000976453 | 0.464792 | 0.115432 | 14 | GGAAGGGGGGGGCAGGG | TGAAAAAGTGGGGT | - |
| abi4#MA0123.1 | -7 | 0.00349157 | 1.66199 | 0.366897 | 10 | GGAAGGGGGGGGCAGGG | GGGGGCACCG | - |
| En1#MA0027.1 | -2 | 0.00401689 | 1.91204 | 0.379888 | 11 | GGAAGGGGGGGGCAGGG | AAGTAGTGCCC | + |
| Pax5#MA0014.1 | 0 | 0.00471533 | 2.2445 | 0.405402 | 17 | GGAAGGGGGGGGCAGGG | GGAGCACTGAAGCGTAACCG | + |
| Egr1#MA0162.1 | -4 | 0.00604315 | 2.87654 | 0.432981 | 11 | GGAAGGGGGGGGCAGGG | TGCGTGGGCGT | + |
| NDT80#MA0343.1 | 3 | 0.00666461 | 3.17235 | 0.432981 | 17 | GGAAGGGGGGGGCAGGG | TTGCGTTTTTGTGGCCGGAAA | - |
| PPARG::RXRA#MA0065.2 | -6 | 0.00700507 | 3.33441 | 0.432981 | 11 | GGAAGGGGGGGGCAGGG | GTAGGGCAAAGGTCA | + |
| ESR1#MA0112.2 | 1 | 0.00704223 | 3.3521 | 0.432981 | 17 | GGAAGGGGGGGGCAGGG | AGGTCAGGGTGACCTGGGCC | - |
| MZF1_5-13#MA0057.1 | -1 | 0.00742687 | 3.53519 | 0.432981 | 10 | GGAAGGGGGGGGCAGGG | GGAGGGGGAA | + |
| CRZ1#MA0285.1 | -8 | 0.00778307 | 3.70474 | 0.432981 | 9 | GGAAGGGGGGGGCAGGG | GTGGCTTAG | - |
| PHD1#MA0355.1 | -7 | 0.0112612 | 5.36031 | 0.591667 | 10 | GGAAGGGGGGGGCAGGG | TGCTGCAGGT | - |
| Tal1::Gata1#MA0140.1 | -4 | 0.0125236 | 5.96124 | 0.623365 | 13 | GGAAGGGGGGGGCAGGG | CTGGTGGGGACAGATAAG | + |
| ESR1#MA0112.2 | -7 | 0.0154663 | 7.36194 | 0.720385 | 10 | GGAAGGGGGGGGCAGGG | GGGGTCACGGTGACCTGG | - |
| CHA4#MA0283.1 | -10 | 0.0167425 | 7.96944 | 0.720385 | 7 | GGAAGGGGGGGGCAGGG | GGCGGAGA | + |
| MIG2#MA0338.1 | -10 | 0.0171142 | 8.14634 | 0.720385 | 7 | GGAAGGGGGGGGCAGGG | TGCGGGG | - |
| Zfx#MA0146.1 | -7 | 0.0182892 | 8.70566 | 0.720385 | 10 | GGAAGGGGGGGGCAGGG | GGGGCCGAGGCCTG | + |
| MIG3#MA0339.1 | -10 | 0.0194117 | 9.23996 | 0.720385 | 7 | GGAAGGGGGGGGCAGGG | TGCGGGG | - |
| hkb#MA0450.1 | -8 | 0.0195543 | 9.30784 | 0.720385 | 9 | GGAAGGGGGGGGCAGGG | GGGGCGTGA | + |
| usp#MA0016.1 | -7 | 0.0198048 | 9.42711 | 0.720385 | 10 | GGAAGGGGGGGGCAGGG | GGGGTCACGG | + |