MacroAPE 1083:12-CGCGGCGGCGGCT
From FANTOM5_SSTAR
Full Name: 12-CGCGGCGGCGGCT,CNhs11959,BestGuess:EGR1_f2_HM09(0.69303244198093),T:6852.0(74.70%),B:27632.3(68.98%),P:1e-33
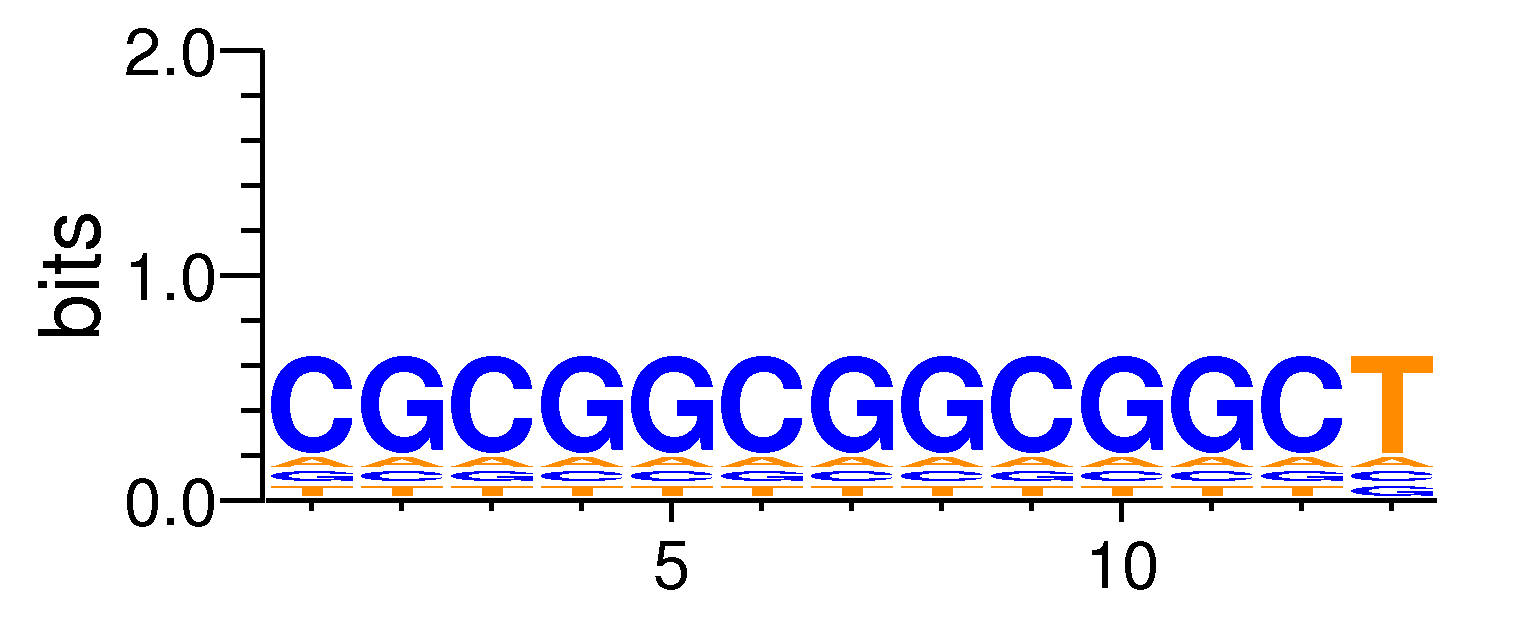
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| STP1#MA0394.1 | 0 | 0.000281542 | 0.134014 | 0.180269 | 8 | CGCGGCGGCGGCT | TGCGGCGC | + |
| abi4#MA0123.1 | 0 | 0.000654162 | 0.311381 | 0.180269 | 10 | CGCGGCGGCGGCT | GGGGGCACCG | - |
| Egr1#MA0162.1 | 0 | 0.000742361 | 0.353364 | 0.180269 | 11 | CGCGGCGGCGGCT | TGCGTGGGCGT | + |
| IME1#MA0320.1 | 0 | 0.00104642 | 0.498098 | 0.180269 | 8 | CGCGGCGGCGGCT | CTCGGCGG | - |
| STP2#MA0395.1 | 6 | 0.00116924 | 0.556556 | 0.180269 | 13 | CGCGGCGGCGGCT | TCGTCGTGCGGCGCCGATCA | - |
| SUT1#MA0399.1 | 0 | 0.00125469 | 0.597231 | 0.180269 | 7 | CGCGGCGGCGGCT | CGCGGGG | + |
| DAL81#MA0290.1 | 3 | 0.00134957 | 0.642396 | 0.180269 | 13 | CGCGGCGGCGGCT | AATCCCGCCCGCGGCTTTT | - |
| NHLH1#MA0048.1 | 1 | 0.00194435 | 0.92551 | 0.227252 | 11 | CGCGGCGGCGGCT | ACGCAGCTGCGC | - |
| Zfx#MA0146.1 | 0 | 0.00417927 | 1.98933 | 0.366842 | 13 | CGCGGCGGCGGCT | GGGGCCGAGGCCTG | + |
| RPN4#MA0373.1 | -3 | 0.00431567 | 2.05426 | 0.366842 | 7 | CGCGGCGGCGGCT | GGTGGCG | + |
| RSC3#MA0374.1 | 0 | 0.00662796 | 3.15491 | 0.381051 | 7 | CGCGGCGGCGGCT | CGCGCGG | + |
| CRZ1#MA0285.1 | -1 | 0.00706205 | 3.36154 | 0.381051 | 9 | CGCGGCGGCGGCT | GTGGCTTAG | - |
| RDS1#MA0361.1 | -4 | 0.00787411 | 3.74808 | 0.381051 | 7 | CGCGGCGGCGGCT | CCGGCCG | - |
| LEU3#MA0324.1 | -1 | 0.00788345 | 3.75252 | 0.381051 | 10 | CGCGGCGGCGGCT | CCGGTAACGG | - |
| Myf#MA0055.1 | -2 | 0.00805393 | 3.83367 | 0.381051 | 11 | CGCGGCGGCGGCT | CAGCAGCTGCTG | + |
| MET31#MA0333.1 | -1 | 0.008698 | 4.14025 | 0.381051 | 9 | CGCGGCGGCGGCT | GGTGTGGCG | + |
| brk#MA0213.1 | -3 | 0.00974088 | 4.63666 | 0.381051 | 8 | CGCGGCGGCGGCT | GGCGCCAG | - |
| btd#MA0443.1 | -2 | 0.00981998 | 4.67431 | 0.381051 | 10 | CGCGGCGGCGGCT | AGGGGGCGGA | + |
| UGA3#MA0410.1 | -5 | 0.0100909 | 4.80328 | 0.381051 | 8 | CGCGGCGGCGGCT | CGGCGGGA | + |
| opa#MA0456.1 | -2 | 0.0101086 | 4.81172 | 0.381051 | 11 | CGCGGCGGCGGCT | CAGCGGGGGGTC | - |
| CHA4#MA0283.1 | -3 | 0.0108263 | 5.1533 | 0.381051 | 8 | CGCGGCGGCGGCT | GGCGGAGA | + |
| TFAP2A#MA0003.1 | 0 | 0.0110119 | 5.24167 | 0.381051 | 9 | CGCGGCGGCGGCT | CCCCCGGGC | - |
| SP1#MA0079.2 | 0 | 0.0111146 | 5.29054 | 0.381051 | 10 | CGCGGCGGCGGCT | GGGGGCGGGG | - |
| UME6#MA0412.1 | -4 | 0.0117576 | 5.59664 | 0.381051 | 9 | CGCGGCGGCGGCT | TCGGCGGCTAATT | + |
| SP1#MA0079.2 | -1 | 0.0118184 | 5.62554 | 0.381051 | 10 | CGCGGCGGCGGCT | GGGGCGGGGT | + |
| Mafb#MA0117.1 | -4 | 0.0137954 | 6.56663 | 0.427101 | 8 | CGCGGCGGCGGCT | GCGTCAGC | - |
| YLL054C#MA0429.1 | -5 | 0.0178695 | 8.50588 | 0.477384 | 7 | CGCGGCGGCGGCT | CGGCCGA | + |