MacroAPE 1083:CNhs10643 bu f2
From FANTOM5_SSTAR
Full Name: CNhs10643_bu_f2
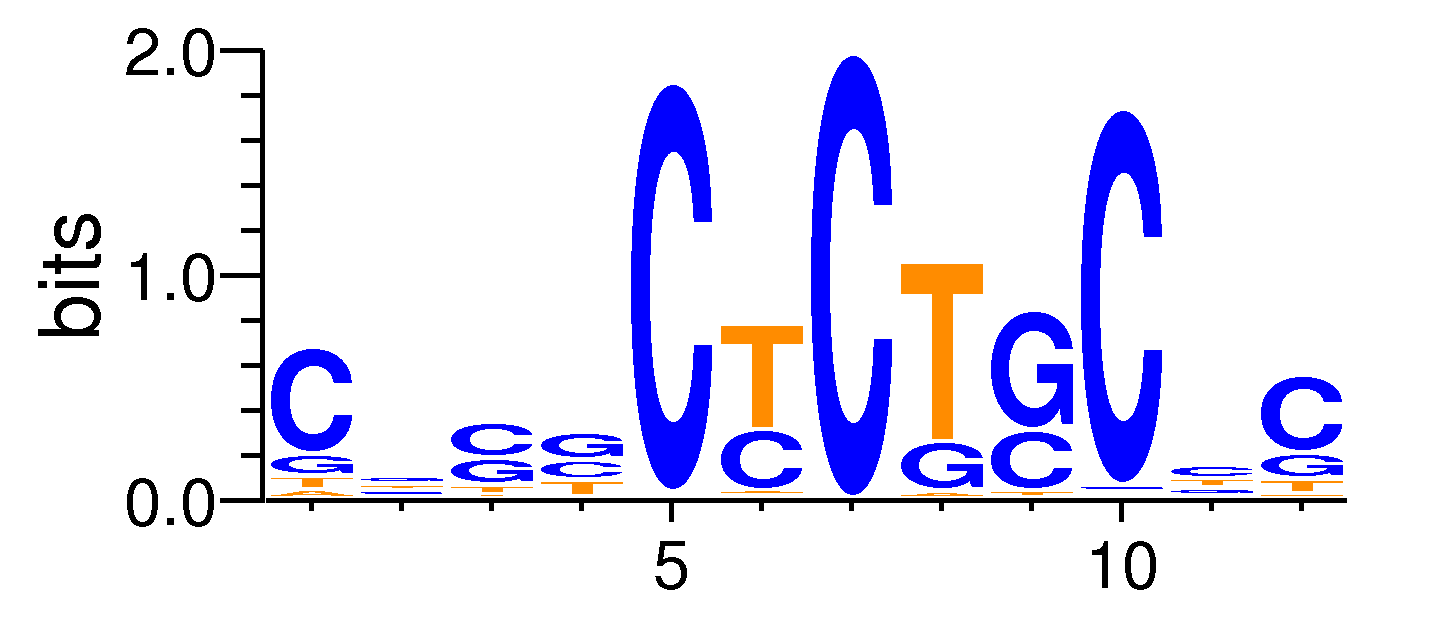
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| Trl#MA0205.1 | -1 | 0.00190118 | 0.90496 | 1 | 10 | CGCGCTCTGCCC | TTGCTCTCTC | + |
| PPARG::RXRA#MA0065.2 | 0 | 0.00396045 | 1.88517 | 1 | 12 | CGCGCTCTGCCC | TGACCTTTGCCCTAC | - |
| SP1#MA0079.2 | -3 | 0.00399996 | 1.90398 | 1 | 9 | CGCGCTCTGCCC | CCCCGCCCCC | + |
| Pax4#MA0068.1 | 18 | 0.00605692 | 2.88309 | 1 | 12 | CGCGCTCTGCCC | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| BAS1#MA0278.1 | 11 | 0.00692273 | 3.29522 | 1 | 10 | CGCGCTCTGCCC | TTGACTTGACTCTGGCTGTGC | - |
| GAL4#MA0299.1 | 1 | 0.00722825 | 3.44065 | 1 | 12 | CGCGCTCTGCCC | GAGTACTGTTCCCCG | - |
| CHA4#MA0283.1 | -3 | 0.00808123 | 3.84667 | 1 | 8 | CGCGCTCTGCCC | TCTCCGCC | - |
| RXR::RAR_DR5#MA0159.1 | 4 | 0.00852912 | 4.05986 | 1 | 12 | CGCGCTCTGCCC | TGACCTCTCCGTGACCT | - |
| HNF4A#MA0114.1 | 0 | 0.0110706 | 5.2696 | 1 | 12 | CGCGCTCTGCCC | TGACCTTTGGCCT | - |
| YGR067C#MA0425.1 | 2 | 0.0115925 | 5.51805 | 1 | 12 | CGCGCTCTGCCC | ACCCCACTTTTTTG | + |
| Zfx#MA0146.1 | 1 | 0.0152827 | 7.27456 | 1 | 12 | CGCGCTCTGCCC | GGGGCCGAGGCCTG | + |
| abi4#MA0123.1 | -4 | 0.0194569 | 9.26148 | 1 | 8 | CGCGCTCTGCCC | CGGTGCCCCC | + |
| IME1#MA0320.1 | -4 | 0.0195898 | 9.32475 | 1 | 8 | CGCGCTCTGCCC | CTCGGCGG | - |
| SUT1#MA0399.1 | -4 | 0.0204243 | 9.72198 | 1 | 7 | CGCGCTCTGCCC | CCCCGCG | - |