MacroAPE 1083:CNhs11331 bl f2
From FANTOM5_SSTAR
Full Name: CNhs11331_bl_f2
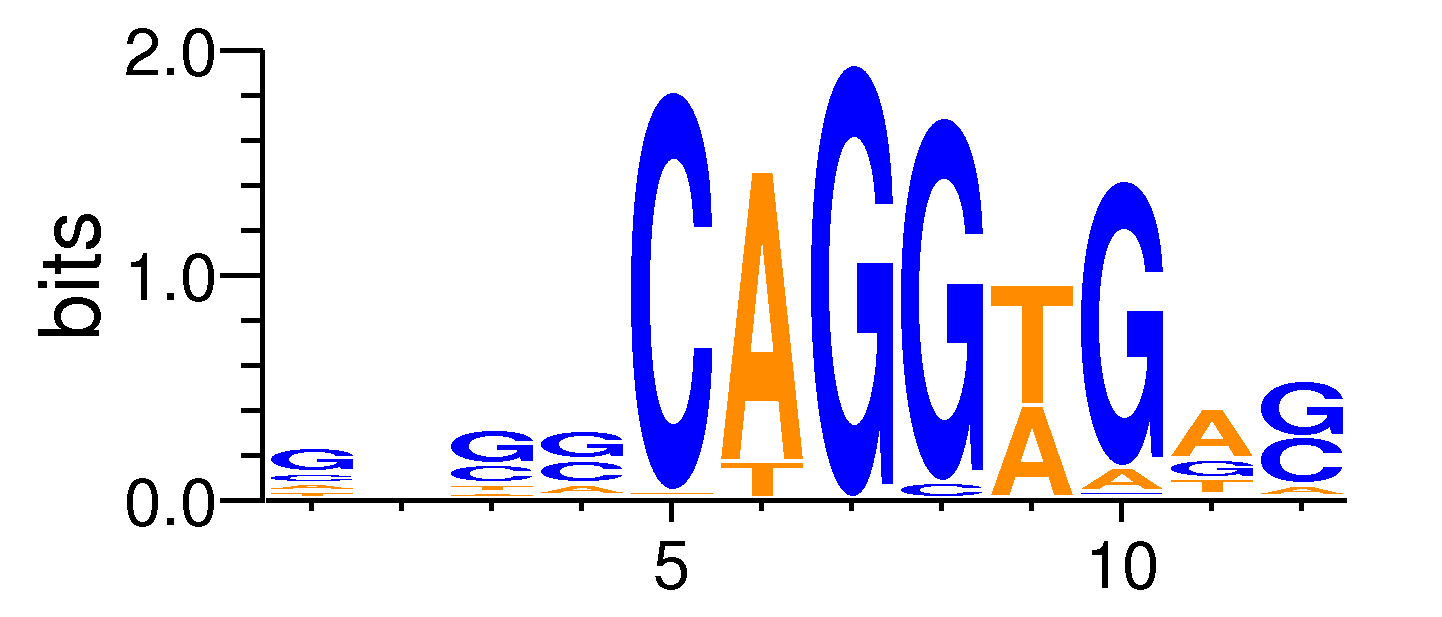
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| sna#MA0086.1 | -4 | 2.56968e-05 | 0.0122317 | 0.0241054 | 6 | GTGGCAGGTGAG | CAGGTG | + |
| ESR1#MA0112.2 | 0 | 7.77009e-05 | 0.0369856 | 0.0364445 | 12 | GTGGCAGGTGAG | GGCCCAGGTCACCCTGACCT | + |
| ZEB1#MA0103.1 | -4 | 0.0002269 | 0.108004 | 0.0709493 | 6 | GTGGCAGGTGAG | CAGGTG | - |
| CBF1#MA0281.1 | -3 | 0.00236961 | 1.12793 | 0.416149 | 8 | GTGGCAGGTGAG | GCACGTGA | + |
| MAX#MA0058.1 | -1 | 0.00269643 | 1.2835 | 0.416149 | 10 | GTGGCAGGTGAG | AACCACGTGA | + |
| TYE7#MA0409.1 | -4 | 0.00284699 | 1.35517 | 0.416149 | 7 | GTGGCAGGTGAG | CACGTGA | + |
| YJL103C#MA0427.1 | 0 | 0.00315137 | 1.50005 | 0.416149 | 11 | GTGGCAGGTGAG | AGCTCCGGAGA | + |
| h#MA0449.1 | -2 | 0.0045675 | 2.17413 | 0.416149 | 10 | GTGGCAGGTGAG | GGCACGTGCC | + |
| USF1#MA0093.1 | -4 | 0.00478437 | 2.27736 | 0.416149 | 7 | GTGGCAGGTGAG | CACGTGG | + |
| Mycn#MA0104.2 | -2 | 0.00541502 | 2.57755 | 0.416149 | 10 | GTGGCAGGTGAG | CGCACGTGGC | + |
| Myc#MA0147.1 | -2 | 0.00640309 | 3.04787 | 0.416149 | 10 | GTGGCAGGTGAG | CGCACGTGGC | + |
| Mycn#MA0104.2 | -4 | 0.00641344 | 3.0528 | 0.416149 | 6 | GTGGCAGGTGAG | CACGTG | - |
| YGR067C#MA0425.1 | 0 | 0.00775303 | 3.69044 | 0.416149 | 12 | GTGGCAGGTGAG | TGAAAAAGTGGGGT | - |
| RXR::RAR_DR5#MA0159.1 | 6 | 0.00816258 | 3.88539 | 0.416149 | 11 | GTGGCAGGTGAG | AGGTCATGGAGAGGTCA | + |
| Arnt#MA0004.1 | -4 | 0.00831976 | 3.9602 | 0.416149 | 6 | GTGGCAGGTGAG | CACGTG | + |
| INO4#MA0322.1 | -3 | 0.0084778 | 4.03543 | 0.416149 | 9 | GTGGCAGGTGAG | GCATGTGAA | + |
| Tal1::Gata1#MA0140.1 | 6 | 0.00959483 | 4.56714 | 0.428602 | 12 | GTGGCAGGTGAG | CTGGTGGGGACAGATAAG | + |
| REI1#MA0364.1 | -3 | 0.0122744 | 5.84262 | 0.458165 | 7 | GTGGCAGGTGAG | TCAGGGG | - |
| ESR1#MA0112.2 | -3 | 0.0124288 | 5.91613 | 0.458165 | 9 | GTGGCAGGTGAG | CCAGGTCACCGTGACCCC | + |
| NHLH1#MA0048.1 | -1 | 0.0130096 | 6.19255 | 0.458165 | 11 | GTGGCAGGTGAG | ACGCAGCTGCGC | - |
| RTG3#MA0376.1 | 3 | 0.0135075 | 6.42956 | 0.458165 | 12 | GTGGCAGGTGAG | ATATTAGCACGTGCTTAGTC | + |
| Esrrb#MA0141.1 | 1 | 0.0158621 | 7.55035 | 0.462005 | 11 | GTGGCAGGTGAG | AGCTCAAGGTCA | + |
| INO2#MA0321.1 | -3 | 0.0160533 | 7.64135 | 0.462005 | 9 | GTGGCAGGTGAG | GCATGTGAA | + |
| Myb#MA0100.1 | -2 | 0.0162527 | 7.73627 | 0.462005 | 8 | GTGGCAGGTGAG | GGCCGTTG | + |
| PHO4#MA0357.1 | -3 | 0.0162527 | 7.73627 | 0.462005 | 8 | GTGGCAGGTGAG | GCACGTGC | + |
| HAC1#MA0310.1 | -2 | 0.0199396 | 9.49126 | 0.535657 | 8 | GTGGCAGGTGAG | GACACGTG | + |