MacroAPE 1083:10-WGACAAC
From FANTOM5_SSTAR
Full Name: 10-WGACAAC,CNhs13239,BestGuess:Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq/Homer(0.741488069271231),T:2239.0(73.77%),B:27365.4(62.56%),P:1e-38
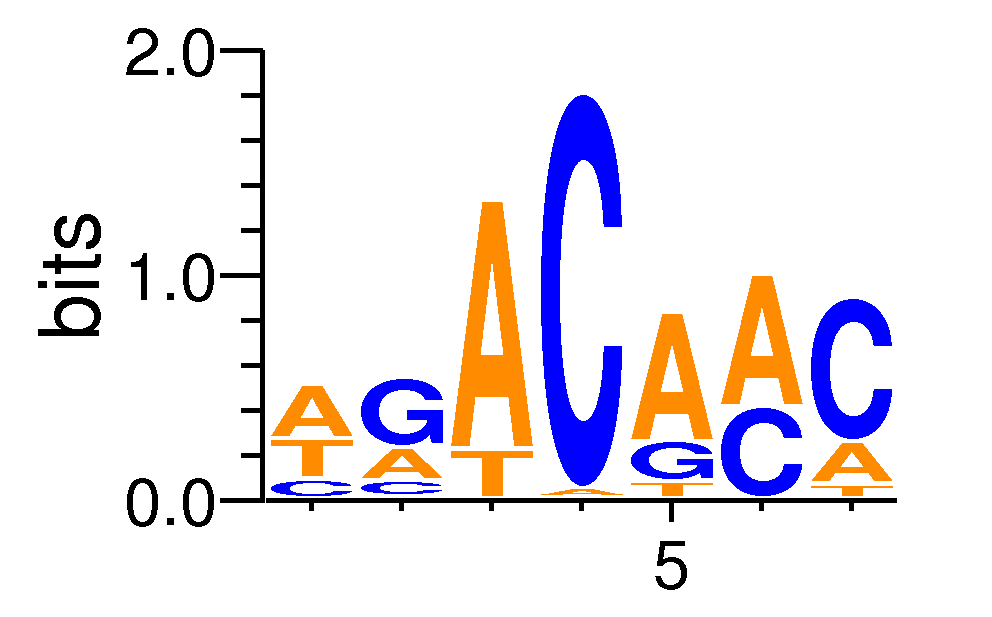
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| HMG-1#MA0044.1 | 2 | 0.00666719 | 3.17358 | 1 | 7 | AGACAAC | GAATACAAC | - |
| Gamyb#MA0034.1 | -1 | 0.00728059 | 3.46556 | 1 | 6 | AGACAAC | GACAACCGCC | + |
| br_Z1#MA0010.1 | 5 | 0.0115413 | 5.49367 | 1 | 7 | AGACAAC | GTAATAAACAAATC | + |
| AFT2#MA0270.1 | -1 | 0.0119311 | 5.67921 | 1 | 6 | AGACAAC | CACACCCC | + |
| BRCA1#MA0133.1 | -2 | 0.0135074 | 6.42952 | 1 | 5 | AGACAAC | ACAACAC | + |
| mirr#MA0233.1 | 0 | 0.0170576 | 8.1194 | 1 | 5 | AGACAAC | AAACA | + |