MacroAPE 1083:COE1 f2
From FANTOM5_SSTAR
Full Name: COE1_f2
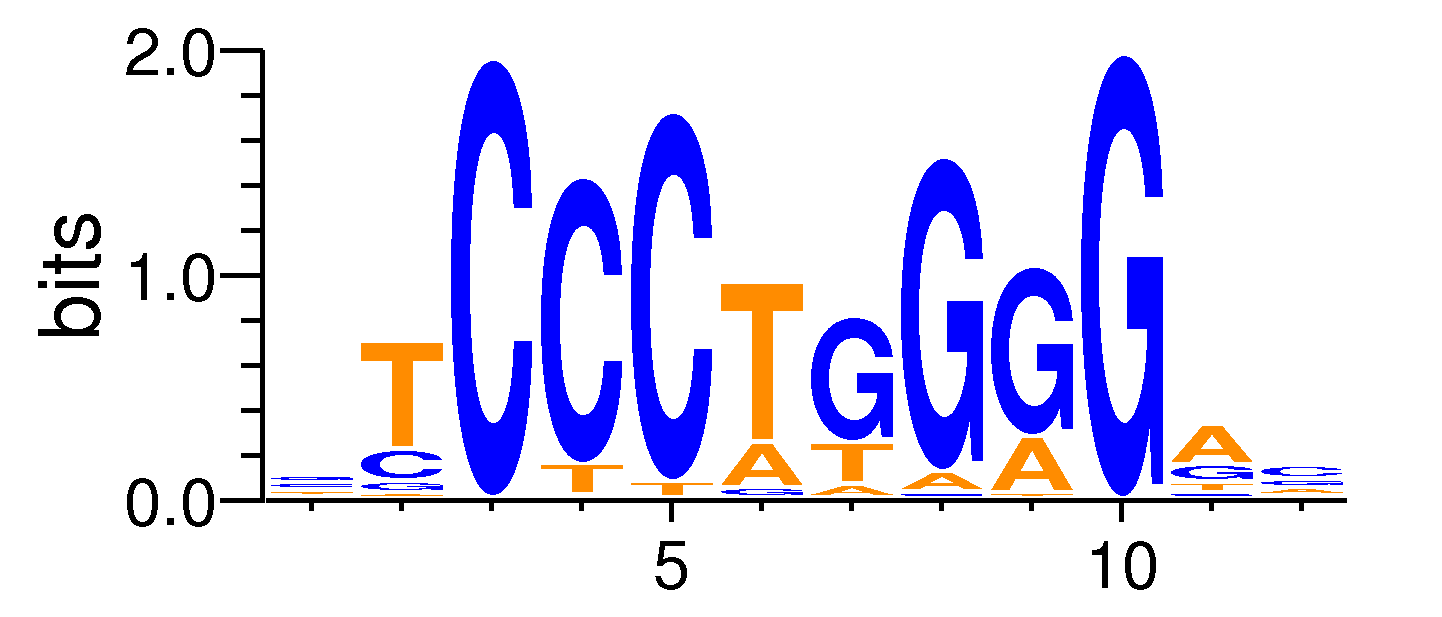
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| EBF1#MA0154.1 | -1 | 1.04188e-06 | 0.000495933 | 0.000986099 | 10 | GTCCCTGGGGAC | TCCCTTGGGT | - |
| Zfp423#MA0116.1 | 2 | 0.00204528 | 0.973553 | 0.540371 | 12 | GTCCCTGGGGAC | GCACCCCTGGGTGCC | - |
| MZF1_1-4#MA0056.1 | -5 | 0.00298846 | 1.42251 | 0.540371 | 6 | GTCCCTGGGGAC | TGGGGA | + |
| PUT3#MA0358.1 | 0 | 0.00324221 | 1.54329 | 0.540371 | 8 | GTCCCTGGGGAC | TTCCCGGG | - |
| INSM1#MA0155.1 | -1 | 0.00342562 | 1.63059 | 0.540371 | 11 | GTCCCTGGGGAC | TGTCAGGGGGCG | + |
| YJL103C#MA0427.1 | -1 | 0.00465366 | 2.21514 | 0.548664 | 11 | GTCCCTGGGGAC | TCTCCGGAGCT | - |
| TFAP2A#MA0003.1 | -1 | 0.00583696 | 2.77839 | 0.548664 | 9 | GTCCCTGGGGAC | GCCCGGGGG | + |
| REI1#MA0364.1 | -3 | 0.0062908 | 2.99442 | 0.548664 | 7 | GTCCCTGGGGAC | TCAGGGG | - |
| PLAG1#MA0163.1 | 0 | 0.00695638 | 3.31124 | 0.548664 | 12 | GTCCCTGGGGAC | CCCCCTTGGGCCCC | - |
| RME1#MA0370.1 | -1 | 0.00998616 | 4.75341 | 0.58481 | 10 | GTCCCTGGGGAC | TTCCTTTGGA | - |
| YPR022C#MA0436.1 | -3 | 0.0107021 | 5.09419 | 0.58481 | 7 | GTCCCTGGGGAC | CGTGGGG | - |
| NHP10#MA0344.1 | -1 | 0.0117066 | 5.57233 | 0.58481 | 8 | GTCCCTGGGGAC | TCCCCGGC | - |
| RGM1#MA0366.1 | -1 | 0.0117399 | 5.58819 | 0.58481 | 5 | GTCCCTGGGGAC | CCCCT | - |
| ADR1#MA0268.1 | -4 | 0.0134987 | 6.42539 | 0.608384 | 7 | GTCCCTGGGGAC | GTGGGGT | - |
| MIG1#MA0337.1 | -4 | 0.0134987 | 6.42539 | 0.608384 | 7 | GTCCCTGGGGAC | GCGGGGG | - |
| MET28#MA0332.1 | -4 | 0.0181997 | 8.66305 | 0.782971 | 6 | GTCCCTGGGGAC | CTGTGG | + |
| CTCF#MA0139.1 | 5 | 0.0204195 | 9.71969 | 0.82355 | 12 | GTCCCTGGGGAC | TAGCGCCCCCTGGTGGCCA | - |