MacroAPE 1083:Motif290
From FANTOM5_SSTAR
Full Name: motif290
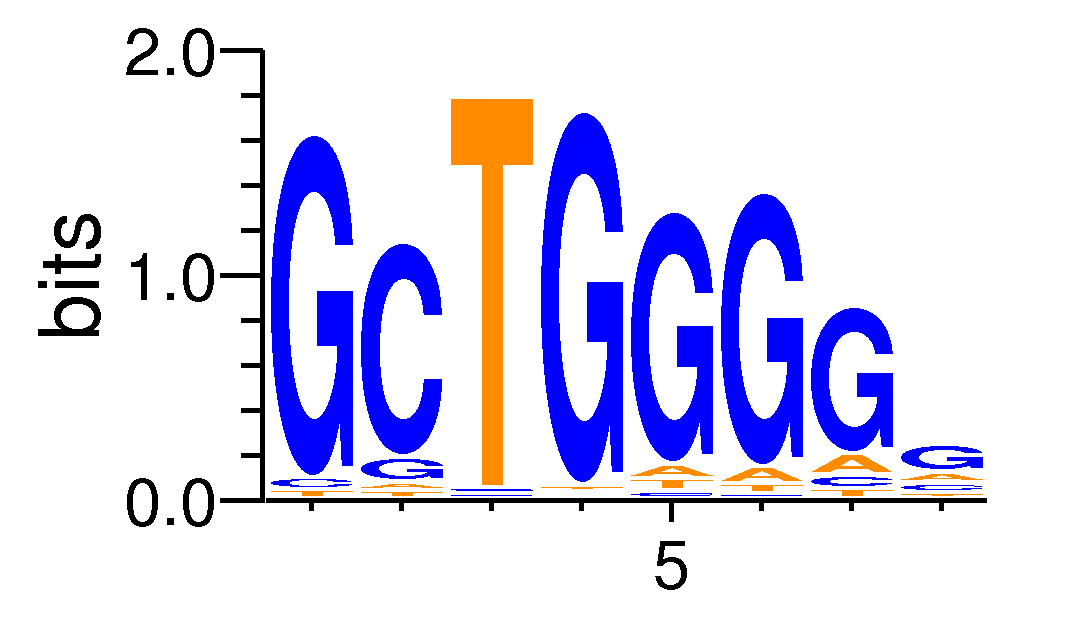
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| opa#MA0456.1 | 2 | 0.000922944 | 0.439321 | 0.361465 | 8 | GCTGGGGG | CAGCGGGGGGTC | - |
| NHP10#MA0344.1 | 0 | 0.00106458 | 0.50674 | 0.361465 | 8 | GCTGGGGG | GCCGGGGA | + |
| MIG1#MA0337.1 | -1 | 0.00114853 | 0.546699 | 0.361465 | 7 | GCTGGGGG | GCGGGGG | - |
| ACE2#MA0267.1 | 1 | 0.00164809 | 0.784491 | 0.389016 | 6 | GCTGGGGG | TGCTGGT | - |
| YPR022C#MA0436.1 | 0 | 0.00256451 | 1.22071 | 0.443709 | 7 | GCTGGGGG | CGTGGGG | - |
| ADR1#MA0268.1 | -1 | 0.00317647 | 1.512 | 0.443709 | 7 | GCTGGGGG | GTGGGGT | - |
| SWI5#MA0402.1 | 1 | 0.0037053 | 1.76372 | 0.443709 | 7 | GCTGGGGG | TGCTGGTT | + |
| INSM1#MA0155.1 | 2 | 0.0037596 | 1.78957 | 0.443709 | 8 | GCTGGGGG | TGTCAGGGGGCG | + |
| EBF1#MA0154.1 | 2 | 0.00510016 | 2.42768 | 0.457293 | 8 | GCTGGGGG | TCCCTTGGGT | - |
| MET28#MA0332.1 | -1 | 0.00514429 | 2.44868 | 0.457293 | 6 | GCTGGGGG | CTGTGG | + |
| RDS2#MA0362.1 | -1 | 0.00537903 | 2.56042 | 0.457293 | 7 | GCTGGGGG | CTCGGGG | + |
| MIG3#MA0339.1 | 0 | 0.00581205 | 2.76653 | 0.457293 | 7 | GCTGGGGG | TGCGGGG | - |
| MIG2#MA0338.1 | 0 | 0.00680865 | 3.24092 | 0.494498 | 7 | GCTGGGGG | TGCGGGG | - |
| Mafb#MA0117.1 | 0 | 0.00838417 | 3.99087 | 0.554752 | 8 | GCTGGGGG | GCTGACGG | + |
| MZF1_1-4#MA0056.1 | -2 | 0.00881341 | 4.19518 | 0.554752 | 6 | GCTGGGGG | TGGGGA | + |
| Klf4#MA0039.2 | 2 | 0.0109347 | 5.20492 | 0.583489 | 8 | GCTGGGGG | TGGGTGGGGC | + |
| MET4#MA0335.1 | 1 | 0.0112286 | 5.3448 | 0.583489 | 7 | GCTGGGGG | AACTGTGG | + |
| NHLH1#MA0048.1 | 5 | 0.0118918 | 5.66051 | 0.583489 | 7 | GCTGGGGG | ACGCAGCTGCGC | - |
| TFAP2A#MA0003.1 | 0 | 0.013142 | 6.25558 | 0.583489 | 8 | GCTGGGGG | CCCCCGGGC | - |
| REI1#MA0364.1 | 0 | 0.0138101 | 6.57359 | 0.583489 | 7 | GCTGGGGG | TCAGGGG | - |
| znf143#MA0088.1 | 9 | 0.0142054 | 6.76177 | 0.583489 | 8 | GCTGGGGG | GCAAGGCATGATGGGAAATC | - |
| Macho-1#MA0118.1 | -1 | 0.0151599 | 7.2161 | 0.583489 | 7 | GCTGGGGG | TGGGGGGTC | + |
| ZMS1#MA0441.1 | 1 | 0.0151599 | 7.2161 | 0.583489 | 8 | GCTGGGGG | TGCGGGGAA | - |
| SUT1#MA0399.1 | 1 | 0.0154499 | 7.35416 | 0.583489 | 6 | GCTGGGGG | CGCGGGG | + |
| RREB1#MA0073.1 | 2 | 0.0166088 | 7.90577 | 0.603129 | 8 | GCTGGGGG | TGGGGGGGGGTGGTTTGGGG | - |
| RPN4#MA0373.1 | 0 | 0.0185638 | 8.83639 | 0.64603 | 7 | GCTGGGGG | GGTGGCG | + |
| YGR067C#MA0425.1 | 6 | 0.0191586 | 9.11949 | 0.64603 | 8 | GCTGGGGG | TGAAAAAGTGGGGT | - |