MacroAPE 1083:6-GTTAACCAATC
From FANTOM5_SSTAR
Full Name: 6-GTTAACCAATC,CNhs12340,BestGuess:NFYA_f1_HM09(0.713739588807101),T:611.0(16.41%),B:4675.3(10.26%),P:1e-30
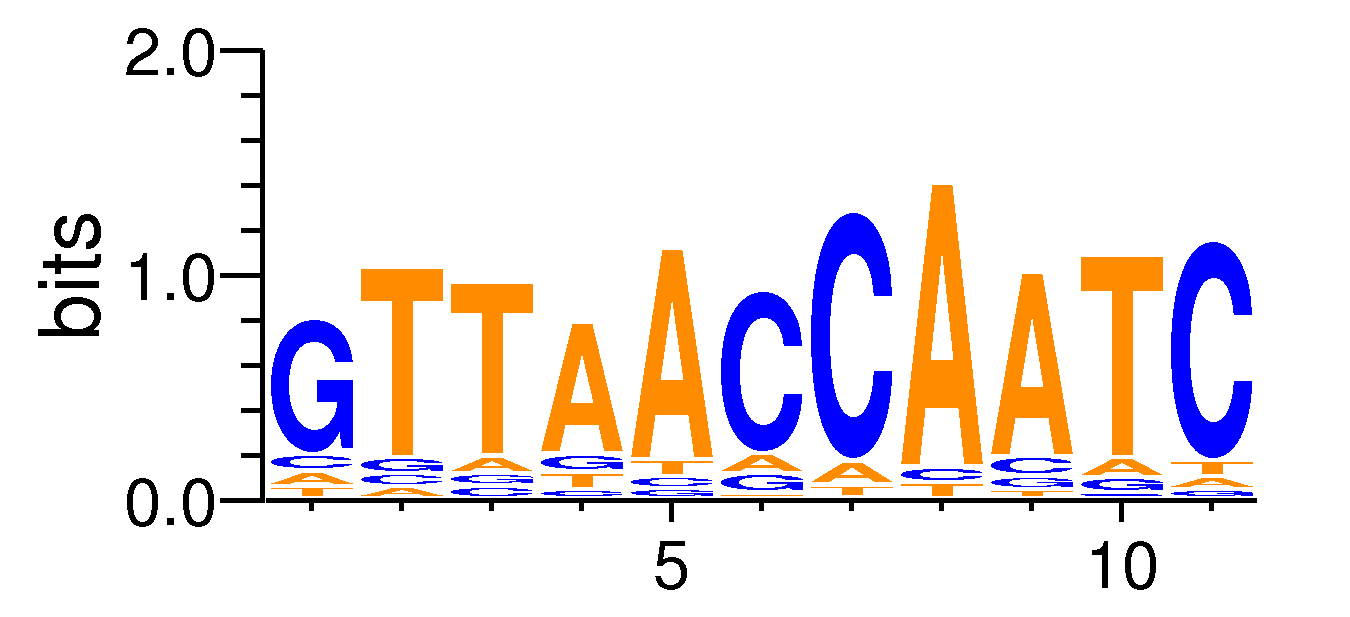
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| NFYA#MA0060.1 | 0 | 0.00025456 | 0.12117 | 0.242279 | 11 | GTTAACCAATC | CTCAGCCAATCAGCGC | + |
| HAP3#MA0314.1 | 1 | 0.000684749 | 0.32594 | 0.325858 | 11 | GTTAACCAATC | TCTGAACCAATCAGA | - |
| SRY#MA0084.1 | -1 | 0.00231658 | 1.10269 | 0.698469 | 9 | GTTAACCAATC | GTAAACAAT | + |
| HLF#MA0043.1 | 1 | 0.00303416 | 1.44426 | 0.698469 | 11 | GTTAACCAATC | GGTTACGCAATT | + |
| HAP2#MA0313.1 | -4 | 0.00366936 | 1.74661 | 0.698469 | 5 | GTTAACCAATC | ACCAA | - |
| fkh#MA0446.1 | -1 | 0.0088151 | 4.19599 | 0.999747 | 10 | GTTAACCAATC | TTAAGCAAACA | - |
| ttx-3::ceh-10#MA0263.1 | -1 | 0.00882297 | 4.19973 | 0.999747 | 10 | GTTAACCAATC | TTAACGAAGCCAAT | - |
| HNF1B#MA0153.1 | 0 | 0.0105324 | 5.0134 | 0.999747 | 11 | GTTAACCAATC | GTTAAATATTAA | - |
| Sox5#MA0087.1 | -3 | 0.0105549 | 5.02413 | 0.999747 | 7 | GTTAACCAATC | AAACAAT | + |
| CIN5#MA0284.1 | -1 | 0.0116703 | 5.55506 | 0.999747 | 10 | GTTAACCAATC | TTACGTAATC | + |
| CEBPA#MA0102.2 | -1 | 0.0127934 | 6.08966 | 0.999747 | 9 | GTTAACCAATC | TTTCGCAAT | + |
| lbe#MA0231.1 | -2 | 0.0134174 | 6.38668 | 0.999747 | 6 | GTTAACCAATC | TAACTA | + |
| Cebpa#MA0102.1 | -1 | 0.0151608 | 7.21655 | 0.999747 | 10 | GTTAACCAATC | TTTCGCAATCTT | + |
| FKH1#MA0296.1 | 8 | 0.0196114 | 9.33502 | 0.999747 | 11 | GTTAACCAATC | TCCTCTTTGTTTACAATTCA | - |
| br_Z1#MA0010.1 | 3 | 0.0196544 | 9.3555 | 0.999747 | 11 | GTTAACCAATC | GTAATAAACAAATC | + |