MacroAPE 1083:3-CCCCCCTTTTTT
From FANTOM5_SSTAR
Full Name: 3-CCCCCCTTTTTT,BestGuess:SEP3(MADS)/Arabidoposis-Flower-Sep3-ChIP-Seq/Homer(0.693294444999093),T:1732.0(31.85%),B:9477.2(22.18%),P:1e-52
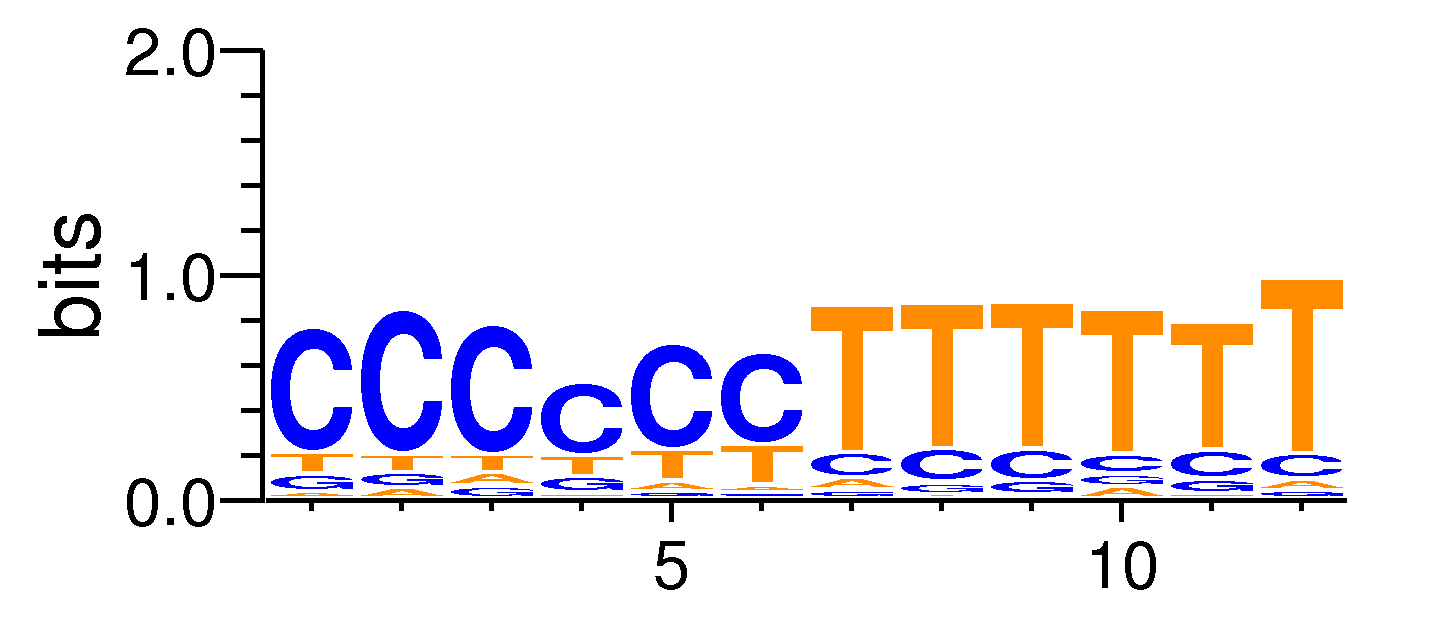
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| Klf4#MA0039.1 | 0 | 0.000713758 | 0.339749 | 0.279391 | 10 | CCCCCCTTTTTT | CCTTCCTTTA | - |
| TBP#MA0108.2 | 1 | 0.000847066 | 0.403204 | 0.279391 | 12 | CCCCCCTTTTTT | CCCCGCCTTTTATAC | - |
| TBP#MA0108.2 | 1 | 0.000882521 | 0.42008 | 0.279391 | 12 | CCCCCCTTTTTT | CCCCGCCTTTTATAC | - |
| YER130C#MA0423.1 | -1 | 0.00125629 | 0.597992 | 0.298289 | 9 | CCCCCCTTTTTT | ACCCCTATT | + |
| YGR067C#MA0425.1 | 1 | 0.00324207 | 1.54323 | 0.49164 | 12 | CCCCCCTTTTTT | ACCCCACTTTTTTG | + |
| FKH1#MA0296.1 | 0 | 0.0033467 | 1.59303 | 0.49164 | 12 | CCCCCCTTTTTT | TCCTCTTTGTTTACAATTCA | - |
| MSN2#MA0341.1 | 0 | 0.00362357 | 1.72482 | 0.49164 | 5 | CCCCCCTTTTTT | CCCCT | - |
| MSN4#MA0342.1 | -2 | 0.00534225 | 2.54291 | 0.563755 | 5 | CCCCCCTTTTTT | CCCCT | - |
| RGM1#MA0366.1 | -2 | 0.00534225 | 2.54291 | 0.563755 | 5 | CCCCCCTTTTTT | CCCCT | - |
| GIS1#MA0306.1 | -1 | 0.00724029 | 3.44638 | 0.687645 | 9 | CCCCCCTTTTTT | ACCCCTAAA | + |
| HMG-I/Y#MA0045.1 | 3 | 0.0129697 | 6.17357 | 0.997635 | 12 | CCCCCCTTTTTT | GTTTTTCCATTTGTTG | - |
| YML081W#MA0431.1 | 1 | 0.0193662 | 9.2183 | 0.997635 | 8 | CCCCCCTTTTTT | ACCCCGCAC | + |