MacroAPE 1083:4-NTGTTTTTTTKTC
From FANTOM5_SSTAR
Full Name: 4-NTGTTTTTTTKTC,CNhs13238,BestGuess:Cdx2(Homeobox)/mES-Cdx2-ChIP-Seq/Homer(0.720601838754395),T:1131.0(43.60%),B:14362.2(32.67%),P:1e-30
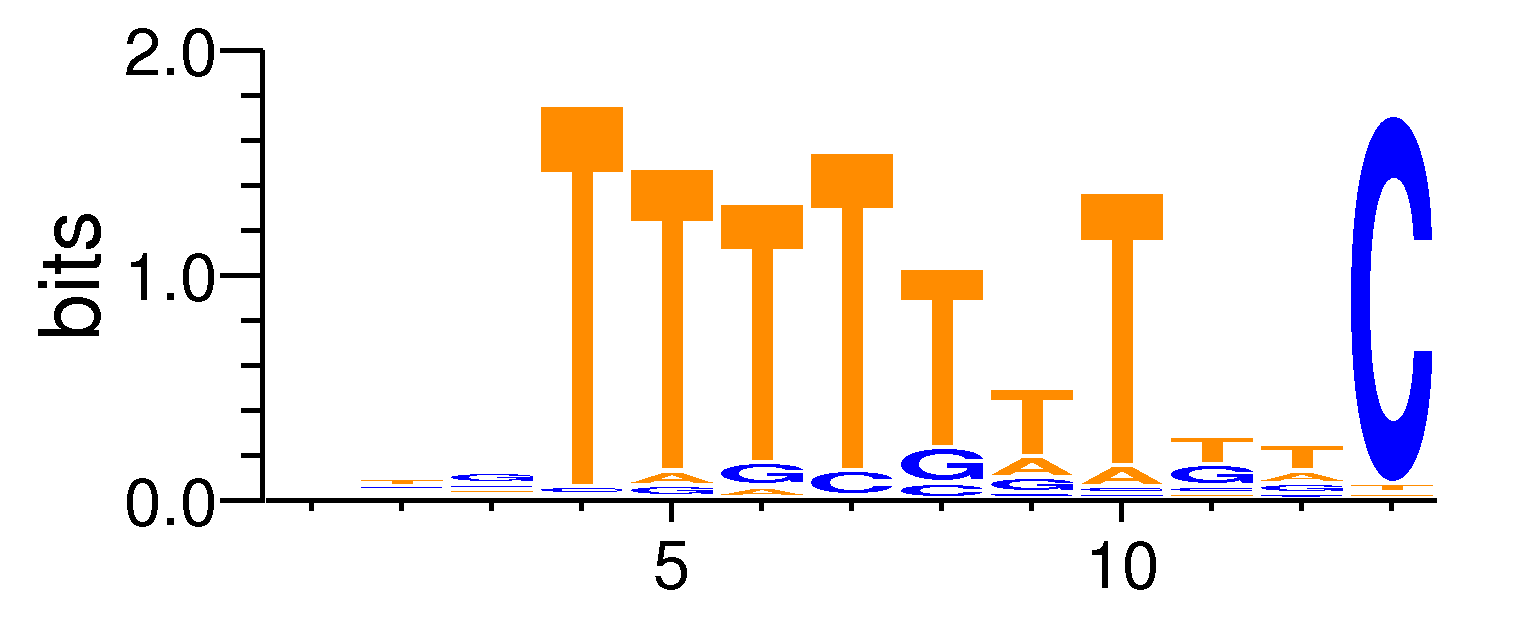
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| hb#MA0049.1 | -3 | 3.35157e-05 | 0.0159535 | 0.0318315 | 10 | ATGTTTTTTTTTC | TTTTTTATGC | - |
| NDT80#MA0343.1 | 2 | 0.000302287 | 0.143889 | 0.143549 | 13 | ATGTTTTTTTTTC | TTGCGTTTTTGTGGCCGGAAA | - |
| AZF1#MA0277.1 | -3 | 0.00217782 | 1.03664 | 0.605295 | 9 | ATGTTTTTTTTTC | TTTCTTTTT | - |
| Foxd3#MA0041.1 | 2 | 0.00254929 | 1.21346 | 0.605295 | 10 | ATGTTTTTTTTTC | GAATGTTTGTTT | + |
| FKH1#MA0296.1 | 1 | 0.00357403 | 1.70124 | 0.673571 | 13 | ATGTTTTTTTTTC | TCCTCTTTGTTTACAATTCA | - |
| STAT1#MA0137.1 | 0 | 0.00425679 | 2.02623 | 0.673571 | 13 | ATGTTTTTTTTTC | CAGTTTCATTTTCC | - |
| FOXI1#MA0042.1 | 2 | 0.00496447 | 2.36309 | 0.673571 | 10 | ATGTTTTTTTTTC | GGATGTTTGTTT | + |
| id1#MA0120.1 | -2 | 0.00682895 | 3.25058 | 0.810724 | 11 | ATGTTTTTTTTTC | TTTTCCTTTTCG | + |
| Evi1#MA0029.1 | -1 | 0.00872093 | 4.15116 | 0.920299 | 12 | ATGTTTTTTTTTC | TGTTATCTTATCTT | - |
| br_Z1#MA0010.1 | 1 | 0.0112764 | 5.36758 | 0.997635 | 13 | ATGTTTTTTTTTC | GATTTGTCTATTAC | - |
| HMG-I/Y#MA0045.1 | 0 | 0.0162873 | 7.75274 | 0.997635 | 13 | ATGTTTTTTTTTC | GTTTTTCCATTTGTTG | - |
| SFP1#MA0378.1 | 7 | 0.0168423 | 8.01691 | 0.997635 | 13 | ATGTTTTTTTTTC | CCGTAGAAAATTTTTTTCAAT | - |
| HCM1#MA0317.1 | -3 | 0.0185912 | 8.84939 | 0.997635 | 8 | ATGTTTTTTTTTC | TTGTTTAT | - |
| br_Z3#MA0012.1 | -1 | 0.0193611 | 9.21589 | 0.997635 | 11 | ATGTTTTTTTTTC | CTTTTAGTTTA | - |