MacroAPE 1083:9-TDDCRNMASM
From FANTOM5_SSTAR
Full Name: 9-TDDCRNMASM,CNhs10860,BestGuess:CEBPB_f1_HM09(0.709901533954884),T:948.0(36.59%),B:11030.9(23.95%),P:1e-46
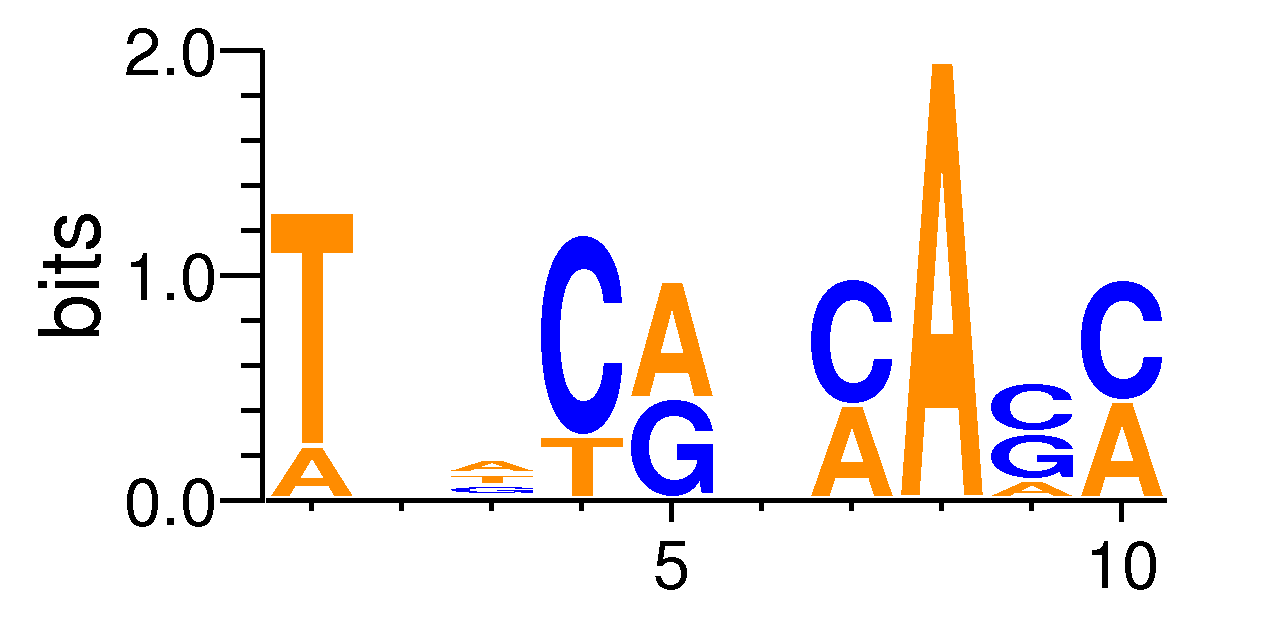
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| CEBPA#MA0102.2 | 1 | 0.000969325 | 0.461399 | 0.395782 | 8 | TTACACCACC | ATTGCGAAA | - |
| HLF#MA0043.1 | 2 | 0.00165485 | 0.787709 | 0.395782 | 10 | TTACACCACC | TATTACGTAACC | - |
| Cebpa#MA0102.1 | 0 | 0.00165485 | 0.787709 | 0.395782 | 10 | TTACACCACC | TTTCGCAATCTT | + |
| BRCA1#MA0133.1 | -2 | 0.00226048 | 1.07599 | 0.395782 | 7 | TTACACCACC | ACAACAC | + |
| Tal1::Gata1#MA0140.1 | 6 | 0.00284909 | 1.35617 | 0.395782 | 10 | TTACACCACC | CTTATCTGTCCCCACCAG | - |
| CIN5#MA0284.1 | 0 | 0.00589603 | 2.80651 | 0.622194 | 10 | TTACACCACC | TTACGTAATC | + |
| Pax4#MA0068.1 | 6 | 0.00672086 | 3.19913 | 0.6364 | 10 | TTACACCACC | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| YAP6#MA0418.1 | 7 | 0.00751203 | 3.57573 | 0.6364 | 10 | TTACACCACC | GGTCTGATTACGTAAGCGAC | + |
| Sox2#MA0143.1 | 1 | 0.00868696 | 4.13499 | 0.6364 | 10 | TTACACCACC | TTTGCATAACAAAGG | - |
| CREB1#MA0018.2 | 0 | 0.0108987 | 5.1878 | 0.738814 | 8 | TTACACCACC | TGACGTCA | - |
| SMP1#MA0383.1 | 10 | 0.0116686 | 5.55424 | 0.738814 | 10 | TTACACCACC | TTACCTATAATTAAATTAGCA | + |
| RPN4#MA0373.1 | -3 | 0.0179596 | 8.54878 | 0.947618 | 7 | TTACACCACC | CGCCACC | - |
| CST6#MA0286.1 | 0 | 0.0208874 | 9.94239 | 0.997635 | 9 | TTACACCACC | TTACGTCAT | - |