MacroAPE 1083:CNhs11853 bu f1
From FANTOM5_SSTAR
Full Name: CNhs11853_bu_f1
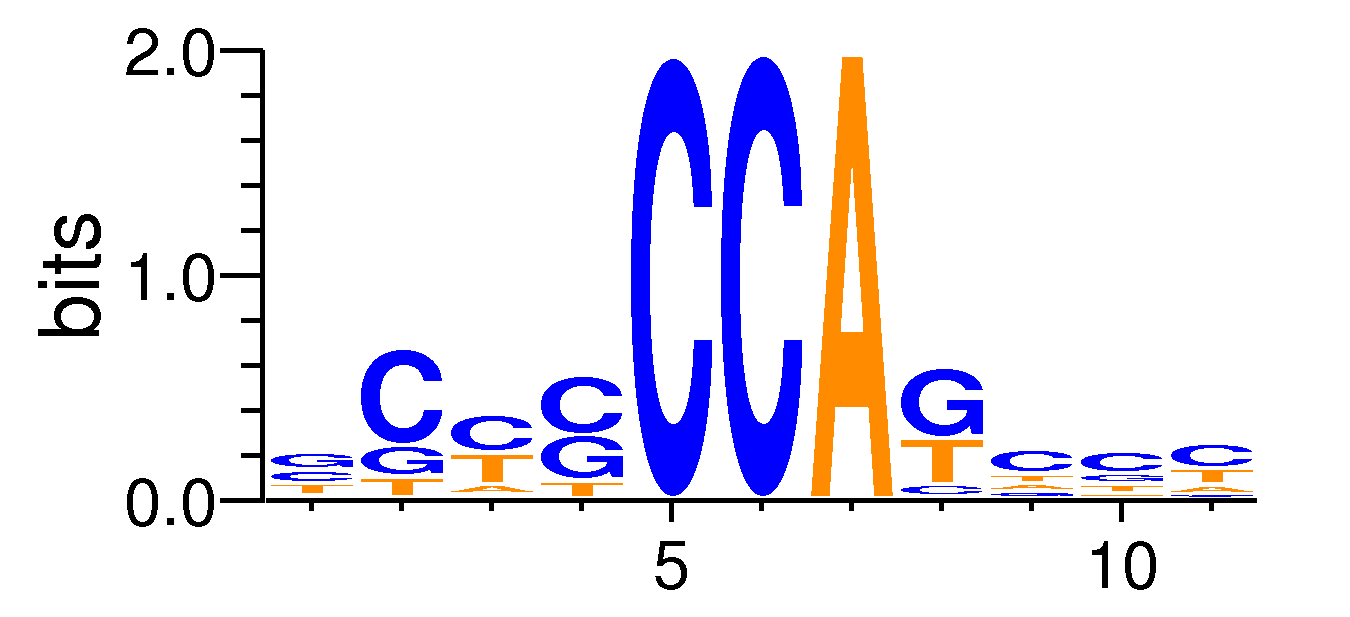
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| znf143#MA0088.1 | 2 | 0.00203359 | 0.967988 | 0.861502 | 11 | GCCCCCAGCCC | GATTTCCCATAATGCCTTGC | + |
| ACE2#MA0267.1 | -3 | 0.0027242 | 1.29672 | 0.861502 | 7 | GCCCCCAGCCC | ACCAGCA | + |
| MZF1_5-13#MA0057.1 | 0 | 0.00372723 | 1.77416 | 0.861502 | 10 | GCCCCCAGCCC | TTCCCCCTAC | - |
| INSM1#MA0155.1 | 1 | 0.00428731 | 2.04076 | 0.861502 | 11 | GCCCCCAGCCC | CGCCCCCTGACA | - |
| Hand1::Tcfe2a#MA0092.1 | -1 | 0.00452584 | 2.1543 | 0.861502 | 10 | GCCCCCAGCCC | ATGCCAGACC | - |
| YY1#MA0095.1 | -3 | 0.00675287 | 3.21437 | 0.890346 | 6 | GCCCCCAGCCC | GCCATC | + |
| SP1#MA0079.2 | -3 | 0.00701254 | 3.33797 | 0.890346 | 8 | GCCCCCAGCCC | CCCCGCCCCC | + |
| Tcfcp2l1#MA0145.1 | -4 | 0.00810151 | 3.85632 | 0.890346 | 7 | GCCCCCAGCCC | CCAGTTCAAACCAG | + |
| MET31#MA0333.1 | -2 | 0.00841927 | 4.00757 | 0.890346 | 9 | GCCCCCAGCCC | CGCCACACT | - |
| Gata1#MA0035.1 | -4 | 0.0102288 | 4.8689 | 0.928348 | 6 | GCCCCCAGCCC | CCATCC | - |
| PHD1#MA0355.1 | 0 | 0.0133812 | 6.36943 | 0.928348 | 10 | GCCCCCAGCCC | ACCTGCAGCA | + |
| Pax4#MA0068.1 | 19 | 0.0136914 | 6.51709 | 0.928348 | 11 | GCCCCCAGCCC | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| SP1#MA0079.2 | -2 | 0.0149711 | 7.12623 | 0.928348 | 9 | GCCCCCAGCCC | ACCCCGCCCC | - |
| YGR067C#MA0425.1 | -1 | 0.015124 | 7.199 | 0.928348 | 10 | GCCCCCAGCCC | ACCCCACTTTTTTG | + |
| Tal1::Gata1#MA0140.1 | 10 | 0.0166581 | 7.92926 | 0.928348 | 8 | GCCCCCAGCCC | CTTATCTGTCCCCACCAG | - |
| ELK1#MA0028.1 | -1 | 0.0176653 | 8.4087 | 0.928348 | 10 | GCCCCCAGCCC | CTTCCGGCTC | - |
| Macho-1#MA0118.1 | 2 | 0.0180327 | 8.58356 | 0.928348 | 7 | GCCCCCAGCCC | GAACCCCCA | - |
| Cebpa#MA0102.1 | 1 | 0.0182272 | 8.67616 | 0.928348 | 11 | GCCCCCAGCCC | TTTCGCAATCTT | + |
| Mafb#MA0117.1 | -1 | 0.0186055 | 8.85621 | 0.928348 | 8 | GCCCCCAGCCC | GCGTCAGC | - |
| YPR022C#MA0436.1 | -2 | 0.0197363 | 9.3945 | 0.928348 | 7 | GCCCCCAGCCC | CCCCACG | + |
| MIG1#MA0337.1 | -1 | 0.0209157 | 9.95586 | 0.928348 | 7 | GCCCCCAGCCC | CCCCCGC | + |