MacroAPE 1083:ZN219 f1
From FANTOM5_SSTAR
Full Name: ZN219_f1
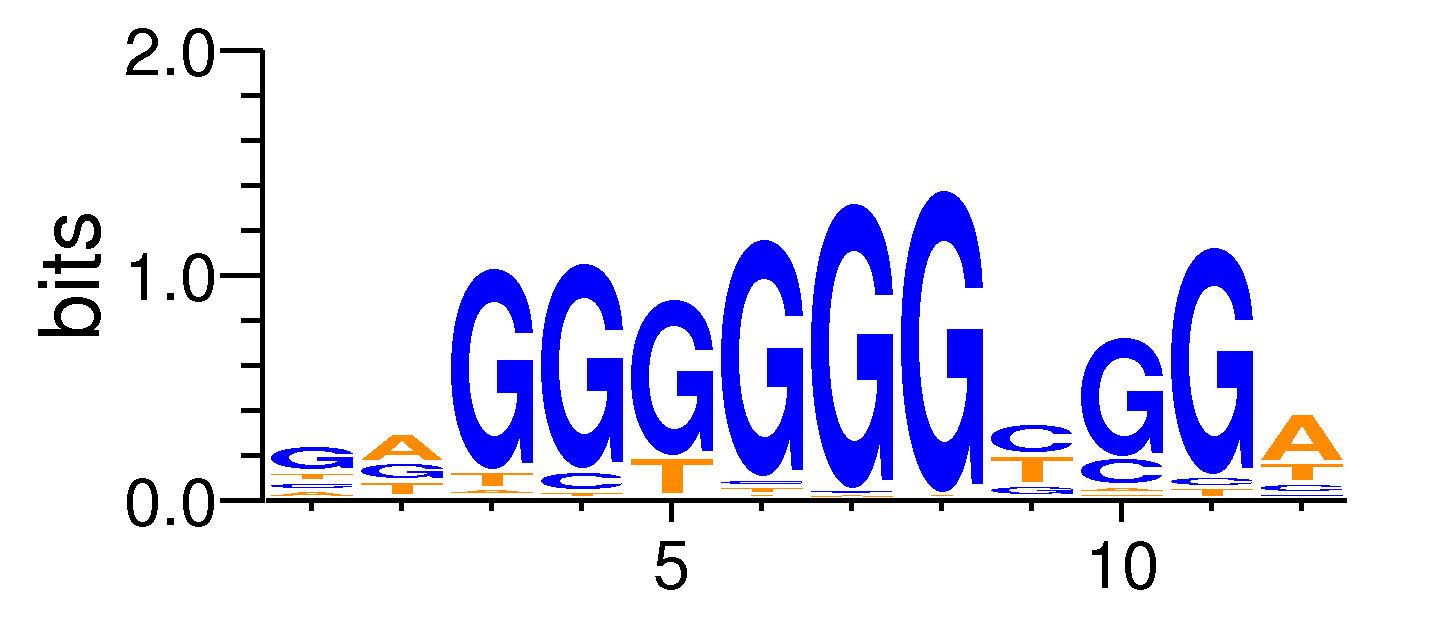
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| btd#MA0443.1 | -2 | 1.31235e-05 | 0.00624681 | 0.0122842 | 10 | GAGGGGGGCGGA | AGGGGGCGGA | + |
| RREB1#MA0073.1 | 2 | 0.000132168 | 0.0629122 | 0.0618575 | 12 | GAGGGGGGCGGA | TGGGGGGGGGTGGTTTGGGG | - |
| SP1#MA0079.2 | -2 | 0.000332983 | 0.1585 | 0.103895 | 10 | GAGGGGGGCGGA | GGGGCGGGGT | + |
| SP1#MA0079.2 | -3 | 0.000631625 | 0.300654 | 0.123282 | 9 | GAGGGGGGCGGA | GGGGGCGGGG | - |
| Macho-1#MA0118.1 | -1 | 0.00065853 | 0.31346 | 0.123282 | 9 | GAGGGGGGCGGA | TGGGGGGTC | + |
| YML081W#MA0431.1 | 0 | 0.000936283 | 0.445671 | 0.146066 | 9 | GAGGGGGGCGGA | GTGCGGGGT | - |
| MIG2#MA0338.1 | -1 | 0.00124179 | 0.59109 | 0.158738 | 7 | GAGGGGGGCGGA | TGCGGGG | - |
| Pax4#MA0068.1 | 2 | 0.00142843 | 0.679934 | 0.158738 | 12 | GAGGGGGGCGGA | GGGGGGGGAGTGGAGTATTGGAAATTTTTC | - |
| AFT2#MA0270.1 | -2 | 0.00152626 | 0.726502 | 0.158738 | 8 | GAGGGGGGCGGA | GGGGTGTG | - |
| MSN2#MA0341.1 | -3 | 0.00213203 | 1.01485 | 0.199567 | 5 | GAGGGGGGCGGA | AGGGG | + |
| MIG3#MA0339.1 | -1 | 0.00309304 | 1.47228 | 0.229083 | 7 | GAGGGGGGCGGA | TGCGGGG | - |
| Klf4#MA0039.2 | -2 | 0.00314426 | 1.49667 | 0.229083 | 10 | GAGGGGGGCGGA | TGGGTGGGGC | + |
| Egr1#MA0162.1 | 0 | 0.00318157 | 1.51443 | 0.229083 | 11 | GAGGGGGGCGGA | TGCGTGGGCGT | + |
| ADR1#MA0268.1 | -2 | 0.0039793 | 1.89415 | 0.248319 | 7 | GAGGGGGGCGGA | GTGGGGT | - |
| MIG1#MA0337.1 | -2 | 0.0039793 | 1.89415 | 0.248319 | 7 | GAGGGGGGCGGA | GCGGGGG | - |
| opa#MA0456.1 | 2 | 0.00436052 | 2.07561 | 0.255102 | 10 | GAGGGGGGCGGA | CAGCGGGGGGTC | - |
| YPR022C#MA0436.1 | -1 | 0.00507494 | 2.41567 | 0.272248 | 7 | GAGGGGGGCGGA | CGTGGGG | - |
| MSN4#MA0342.1 | -1 | 0.00523531 | 2.49201 | 0.272248 | 5 | GAGGGGGGCGGA | AGGGG | + |
| RPN4#MA0373.1 | -3 | 0.00571823 | 2.72188 | 0.277532 | 7 | GAGGGGGGCGGA | GGTGGCG | + |
| abi4#MA0123.1 | 0 | 0.00592991 | 2.82264 | 0.277532 | 10 | GAGGGGGGCGGA | GGGGGCACCG | - |
| hkb#MA0450.1 | -2 | 0.00682256 | 3.24754 | 0.304105 | 9 | GAGGGGGGCGGA | GGGGCGTGA | + |
| RGM1#MA0366.1 | -1 | 0.00810177 | 3.85644 | 0.344709 | 5 | GAGGGGGGCGGA | AGGGG | + |
| YGR067C#MA0425.1 | 5 | 0.00915039 | 4.35559 | 0.372397 | 9 | GAGGGGGGCGGA | TGAAAAAGTGGGGT | - |
| ZMS1#MA0441.1 | -1 | 0.0100558 | 4.78658 | 0.392195 | 9 | GAGGGGGGCGGA | TGCGGGGAA | - |
| Tal1::Gata1#MA0140.1 | 0 | 0.0110257 | 5.24824 | 0.412821 | 12 | GAGGGGGGCGGA | CTGGTGGGGACAGATAAG | + |
| CRZ1#MA0285.1 | -2 | 0.0127256 | 6.05739 | 0.430099 | 9 | GAGGGGGGCGGA | GTGGCTTAG | - |
| CTCF#MA0139.1 | 6 | 0.012817 | 6.10089 | 0.430099 | 12 | GAGGGGGGCGGA | TGGCCACCAGGGGGCGCTA | + |
| AFT1#MA0269.1 | 1 | 0.0128657 | 6.12405 | 0.430099 | 12 | GAGGGGGGCGGA | CCAATCGGGTGCAATATGTAA | - |
| MZF1_5-13#MA0057.1 | 1 | 0.0135961 | 6.47174 | 0.438845 | 9 | GAGGGGGGCGGA | GGAGGGGGAA | + |
| MZF1_1-4#MA0056.1 | -2 | 0.0149811 | 7.13102 | 0.467432 | 6 | GAGGGGGGCGGA | TGGGGA | + |
| INSM1#MA0155.1 | 2 | 0.0195814 | 9.32073 | 0.591256 | 10 | GAGGGGGGCGGA | TGTCAGGGGGCG | + |
| MET31#MA0333.1 | -1 | 0.0209631 | 9.97844 | 0.613198 | 9 | GAGGGGGGCGGA | GGTGTGGCG | + |