MacroAPE 1083:Motif494
From FANTOM5_SSTAR
Full Name: motif494
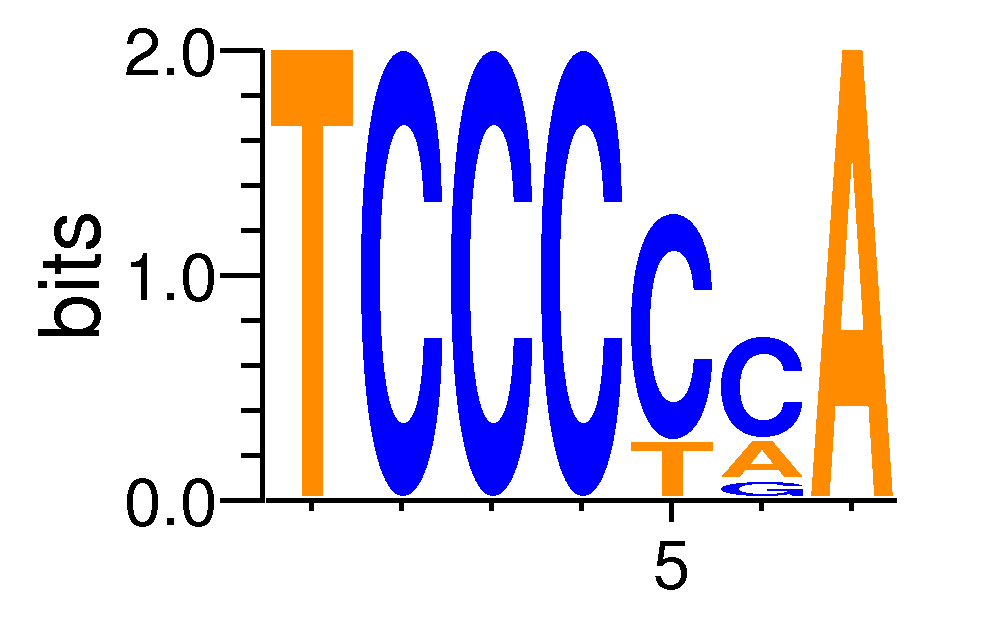
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| MZF1_1-4#MA0056.1 | 0 | 0.000170517 | 0.0811662 | 0.162332 | 6 | TCCCCCA | TCCCCA | - |
| ADR1#MA0268.1 | -1 | 0.00452758 | 2.15513 | 0.881192 | 6 | TCCCCCA | ACCCCAC | + |
| Macho-1#MA0118.1 | 2 | 0.00468455 | 2.22985 | 0.881192 | 7 | TCCCCCA | GAACCCCCA | - |
| MIG1#MA0337.1 | -1 | 0.00496273 | 2.36226 | 0.881192 | 6 | TCCCCCA | CCCCCGC | + |
| MSN2#MA0341.1 | -1 | 0.00597459 | 2.84391 | 0.881192 | 5 | TCCCCCA | CCCCT | - |
| MZF1_5-13#MA0057.1 | 1 | 0.00611423 | 2.91037 | 0.881192 | 7 | TCCCCCA | TTCCCCCTAC | - |
| SP1#MA0079.2 | 4 | 0.00669971 | 3.18906 | 0.881192 | 6 | TCCCCCA | CCCCGCCCCC | + |
| RDS2#MA0362.1 | -1 | 0.00808923 | 3.85047 | 0.881192 | 6 | TCCCCCA | CCCCGAG | - |
| OAF1#MA0348.1 | 2 | 0.0100477 | 4.7827 | 0.881192 | 7 | TCCCCCA | TATCTCCGA | - |
| PPARG::RXRA#MA0065.2 | 7 | 0.0126439 | 6.01847 | 0.881192 | 7 | TCCCCCA | TGACCTTTGCCCTAC | - |
| YPR022C#MA0436.1 | -2 | 0.0141098 | 6.71626 | 0.881192 | 5 | TCCCCCA | CCCCACG | + |
| EBF1#MA0154.1 | -1 | 0.0141433 | 6.7322 | 0.881192 | 6 | TCCCCCA | ACCCAAGGGA | + |
| Su(H)#MA0085.1 | 8 | 0.0146492 | 6.97304 | 0.881192 | 7 | TCCCCCA | ATCTCGGTTCCCACAA | - |
| RREB1#MA0073.1 | -1 | 0.0146794 | 6.98741 | 0.881192 | 6 | TCCCCCA | CCCCAAACCACCCCCCCCCC | + |
| RPH1#MA0372.1 | 0 | 0.017151 | 8.16389 | 0.881192 | 7 | TCCCCCA | ACCCCTAA | + |
| NHP10#MA0344.1 | 0 | 0.0178566 | 8.49975 | 0.881192 | 7 | TCCCCCA | TCCCCGGC | - |
| MSN4#MA0342.1 | -1 | 0.0184451 | 8.77989 | 0.881192 | 5 | TCCCCCA | CCCCT | - |
| GIS1#MA0306.1 | 0 | 0.018807 | 8.95212 | 0.881192 | 7 | TCCCCCA | ACCCCTAAA | + |
| ZMS1#MA0441.1 | 0 | 0.018807 | 8.95212 | 0.881192 | 7 | TCCCCCA | TTCCCCGCA | + |
| USV1#MA0413.1 | 4 | 0.0194029 | 9.23576 | 0.881192 | 7 | TCCCCCA | AAATTCCCCCTGAATTTGTG | + |
| INSM1#MA0155.1 | 1 | 0.01967 | 9.36293 | 0.881192 | 7 | TCCCCCA | CGCCCCCTGACA | - |
| NFKB1#MA0105.1 | 6 | 0.0204371 | 9.72806 | 0.881192 | 5 | TCCCCCA | GGGGAATCCCC | - |