MacroAPE 1083:6-CTTCCTGTCAGAG
From FANTOM5_SSTAR
Full Name: 6-CTTCCTGTCAGAG,CNhs11673,BestGuess:Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq/Homer(0.742504427802236),T:1504.0(44.21%),B:16183.8(35.07%),P:1e-27
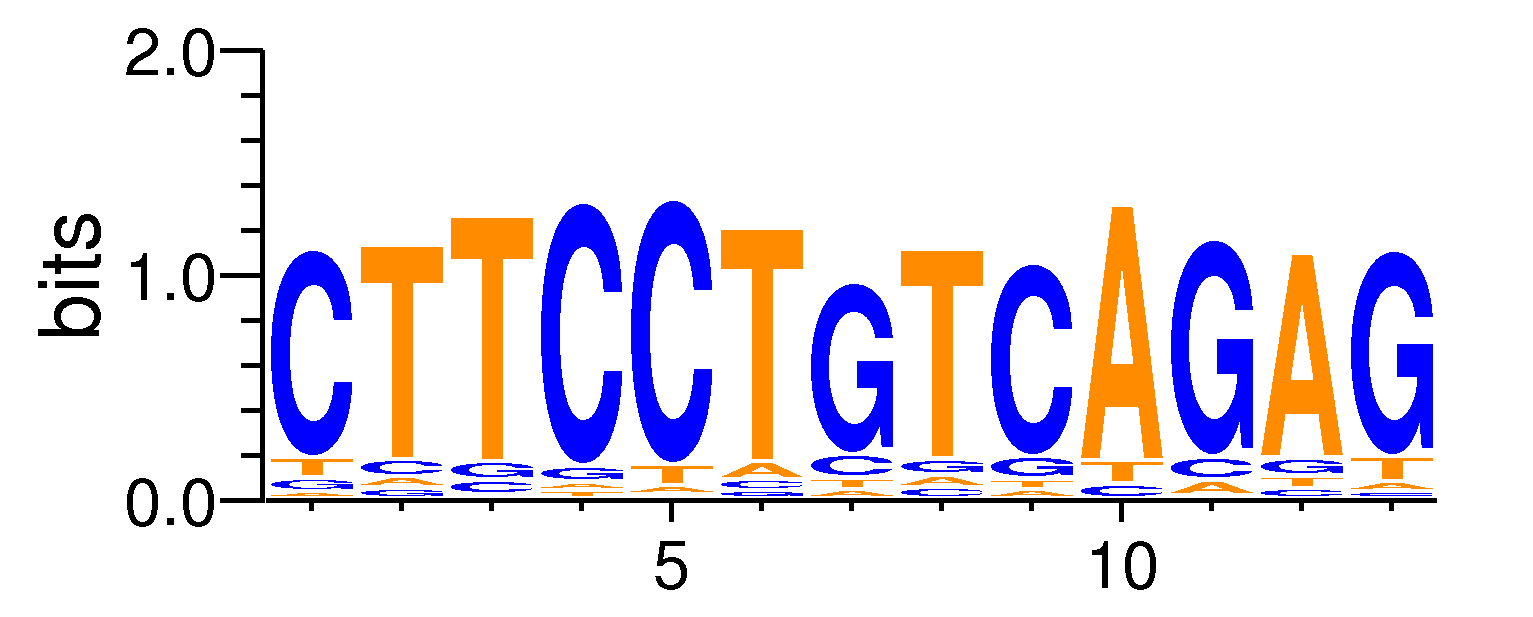
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| achi#MA0207.1 | -4 | 0.000280103 | 0.133329 | 0.169384 | 6 | CTTCCTGTCAGAG | CTGTCA | - |
| hth#MA0227.1 | -4 | 0.000588429 | 0.280092 | 0.169384 | 6 | CTTCCTGTCAGAG | CTGTCA | - |
| vis#MA0252.1 | -4 | 0.000689209 | 0.328063 | 0.169384 | 6 | CTTCCTGTCAGAG | CTGTCA | - |
| Stat3#MA0144.1 | 0 | 0.000716419 | 0.341015 | 0.169384 | 10 | CTTCCTGTCAGAG | CTTCCTGGAA | - |
| Eip74EF#MA0026.1 | 0 | 0.00099104 | 0.471735 | 0.187451 | 7 | CTTCCTGTCAGAG | CTTCCGG | - |
| FEV#MA0156.1 | 1 | 0.00153292 | 0.729669 | 0.223367 | 7 | CTTCCTGTCAGAG | ATTTCCTG | - |
| SPIB#MA0081.1 | -1 | 0.00183164 | 0.871859 | 0.223367 | 7 | CTTCCTGTCAGAG | TTCCTCT | - |
| SPI1#MA0080.2 | 1 | 0.00201171 | 0.957575 | 0.223367 | 6 | CTTCCTGTCAGAG | ACTTCCT | - |
| TOS8#MA0408.1 | -4 | 0.00212567 | 1.01182 | 0.223367 | 8 | CTTCCTGTCAGAG | CTGTCAAA | + |
| SPI1#MA0080.2 | 0 | 0.00430709 | 2.05018 | 0.403509 | 6 | CTTCCTGTCAGAG | CTTCCG | - |
| ELK4#MA0076.1 | 1 | 0.00469331 | 2.23402 | 0.403509 | 8 | CTTCCTGTCAGAG | ACTTCCGGT | - |
| ETS1#MA0098.1 | 0 | 0.00737926 | 3.51253 | 0.525375 | 6 | CTTCCTGTCAGAG | CTTCCG | + |
| GABPA#MA0062.1 | 2 | 0.0076195 | 3.62688 | 0.525375 | 8 | CTTCCTGTCAGAG | CTCTTCCGGT | - |
| GCR2#MA0305.1 | 1 | 0.00777734 | 3.70201 | 0.525375 | 6 | CTTCCTGTCAGAG | GCTTCCT | + |
| STAT1#MA0137.2 | 2 | 0.00895117 | 4.26076 | 0.564358 | 13 | CTTCCTGTCAGAG | GGTTTCCGGGAAATG | - |
| RME1#MA0370.1 | -1 | 0.0102135 | 4.86161 | 0.568187 | 10 | CTTCCTGTCAGAG | TTCCTTTGGA | - |
| exd#MA0222.1 | -5 | 0.0117956 | 5.61472 | 0.596511 | 8 | CTTCCTGTCAGAG | TGTCAAAA | - |
| ELK1#MA0028.1 | 0 | 0.0119841 | 5.70443 | 0.596511 | 10 | CTTCCTGTCAGAG | CTTCCGGCTC | - |