MacroAPE 1083:CCCCCTCTCTCC,motif580
From FANTOM5_SSTAR
Full Name: CCCCCTCTCTCC,motif580,CNhs11772,T:482.0(13.94%),B:5609.6(8.05%),P:1e-30
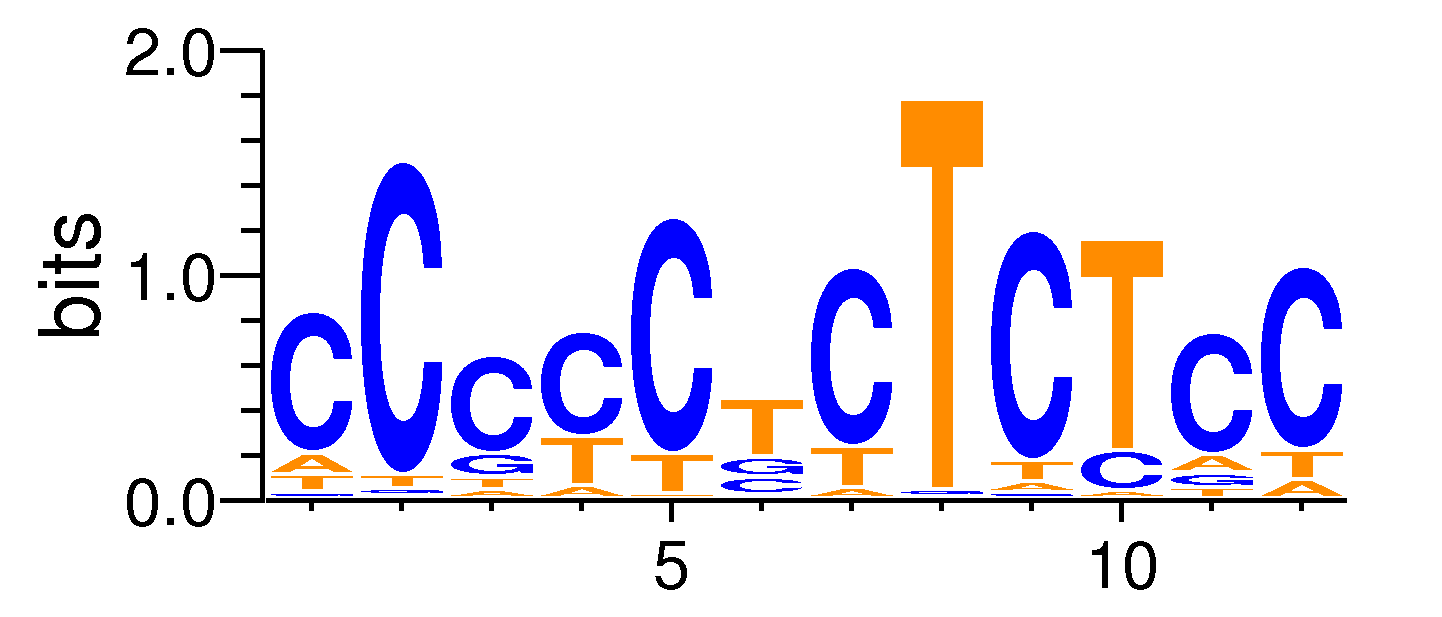
| ExpandMotif matrix |
|---|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| Trl#MA0205.1 | -1 | 0.000256972 | 0.122319 | 0.244576 | 10 | CCCCCTCTCTCC | TTGCTCTCTC | + |
| YML081W#MA0431.1 | 0 | 0.000607954 | 0.289386 | 0.246508 | 9 | CCCCCTCTCTCC | ACCCCGCAC | + |
| SP1#MA0079.2 | -1 | 0.000777009 | 0.369856 | 0.246508 | 10 | CCCCCTCTCTCC | CCCCGCCCCC | + |
| YGR067C#MA0425.1 | 0 | 0.00130712 | 0.622191 | 0.311017 | 12 | CCCCCTCTCTCC | ACCCCACTTTTTTG | + |
| YER130C#MA0423.1 | 0 | 0.00390641 | 1.85945 | 0.743593 | 9 | CCCCCTCTCTCC | ACCCCTATT | + |
| Pax4#MA0068.1 | 18 | 0.00541071 | 2.5755 | 0.858281 | 12 | CCCCCTCTCTCC | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| MSN2#MA0341.1 | -1 | 0.00840755 | 4.00199 | 0.973996 | 5 | CCCCCTCTCTCC | CCCCT | - |
| MSN4#MA0342.1 | -1 | 0.00896011 | 4.26501 | 0.973996 | 5 | CCCCCTCTCTCC | CCCCT | - |
| Gata1#MA0035.2 | -1 | 0.00984799 | 4.68764 | 0.973996 | 11 | CCCCCTCTCTCC | TTCTTATCTGT | - |
| PLAG1#MA0163.1 | 0 | 0.0102336 | 4.87121 | 0.973996 | 12 | CCCCCTCTCTCC | CCCCCTTGGGCCCC | - |
| RGM1#MA0366.1 | -1 | 0.0114759 | 5.46253 | 0.976797 | 5 | CCCCCTCTCTCC | CCCCT | - |
| Klf4#MA0039.1 | -1 | 0.0130165 | 6.19585 | 0.976797 | 10 | CCCCCTCTCTCC | CCTTCCTTTA | - |
| opa#MA0456.1 | 1 | 0.0136991 | 6.52079 | 0.976797 | 11 | CCCCCTCTCTCC | GACCCCCCGCTG | + |
| MIG1#MA0337.1 | 0 | 0.0143683 | 6.83931 | 0.976797 | 7 | CCCCCTCTCTCC | CCCCCGC | + |
| PPARG::RXRA#MA0065.2 | 0 | 0.0194175 | 9.24274 | 0.999747 | 12 | CCCCCTCTCTCC | TGACCTTTGCCCTAC | - |