MacroAPE 1083:EOMES f1
From FANTOM5_SSTAR
Full Name: EOMES_f1
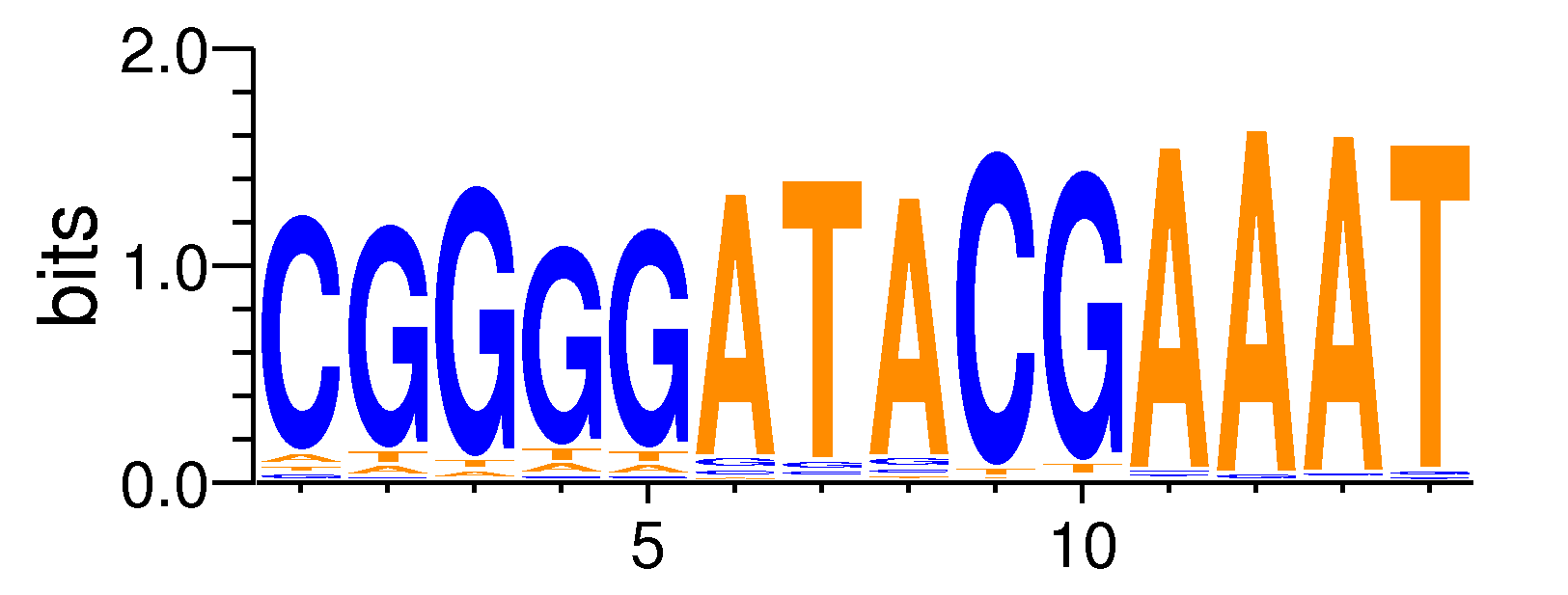
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| OAF1#MA0348.1 | 1 | 0.00076071 | 0.362098 | 0.422698 | 8 | CGGGGATACGAAAT | TCGGAGATA | + |
| YAP3#MA0416.1 | -6 | 0.00217305 | 1.03437 | 0.422698 | 8 | CGGGGATACGAAAT | TACGTAAT | - |
| UPC2#MA0411.1 | -4 | 0.00229818 | 1.09393 | 0.422698 | 7 | CGGGGATACGAAAT | TATACGA | + |
| STB5#MA0392.1 | 0 | 0.00246799 | 1.17476 | 0.422698 | 8 | CGGGGATACGAAAT | CGGTGTTA | + |
| MZF1_1-4#MA0056.1 | 0 | 0.00249782 | 1.18896 | 0.422698 | 6 | CGGGGATACGAAAT | TGGGGA | + |
| HAP1#MA0312.1 | 0 | 0.00268251 | 1.27687 | 0.422698 | 8 | CGGGGATACGAAAT | CGGAGATA | + |
| HLF#MA0043.1 | -3 | 0.00405777 | 1.9315 | 0.509666 | 11 | CGGGGATACGAAAT | GGTTACGCAATT | + |
| YRR1#MA0439.1 | 1 | 0.00431256 | 2.05278 | 0.509666 | 10 | CGGGGATACGAAAT | GCGGAGATAAG | - |
| PEND#MA0127.1 | -4 | 0.00507168 | 2.41412 | 0.532782 | 10 | CGGGGATACGAAAT | AATAAGAAGT | - |
| GSM1#MA0308.1 | 7 | 0.00629369 | 2.9958 | 0.577291 | 14 | CGGGGATACGAAAT | TATACTCCGGAGTTTTTTAAT | - |
| YDR520C#MA0422.1 | 1 | 0.00793267 | 3.77595 | 0.577291 | 9 | CGGGGATACGAAAT | CCGGAGATAA | + |
| STAT1#MA0137.1 | -2 | 0.00799272 | 3.80453 | 0.577291 | 12 | CGGGGATACGAAAT | GGAAAACGAAACTG | + |
| Su(H)#MA0085.1 | 2 | 0.00854947 | 4.06955 | 0.577291 | 14 | CGGGGATACGAAAT | CTGTGGGAAACGAGAT | + |
| YPR196W#MA0437.1 | 0 | 0.00859858 | 4.09292 | 0.577291 | 9 | CGGGGATACGAAAT | CGGAGAAAT | - |
| gt#MA0447.1 | -4 | 0.0098548 | 4.69088 | 0.577291 | 10 | CGGGGATACGAAAT | ATTACGTAAT | + |
| NHP10#MA0344.1 | 2 | 0.0103801 | 4.94095 | 0.577291 | 6 | CGGGGATACGAAAT | GCCGGGGA | + |
| usp#MA0016.1 | -1 | 0.0133476 | 6.35345 | 0.701086 | 10 | CGGGGATACGAAAT | GGGGTCACGG | + |
| SNT2#MA0384.1 | 0 | 0.014212 | 6.76492 | 0.707201 | 11 | CGGGGATACGAAAT | CGGCGCTACCA | - |
| PDR8#MA0354.1 | 1 | 0.0156353 | 7.44241 | 0.739124 | 7 | CGGGGATACGAAAT | GCGGAGAT | + |
| Gata1#MA0035.2 | -1 | 0.0182397 | 8.68208 | 0.815725 | 11 | CGGGGATACGAAAT | ACAGATAAGAA | + |
| GATA2#MA0036.1 | -3 | 0.0205066 | 9.76114 | 0.815725 | 5 | CGGGGATACGAAAT | GGATA | + |
| YAP1#MA0415.1 | 1 | 0.020621 | 9.81558 | 0.815725 | 14 | CGGGGATACGAAAT | ACGAGCTTACGTAAGCAAAT | - |