MacroAPE 1083:HXA13 f1
From FANTOM5_SSTAR
Full Name: HXA13_f1
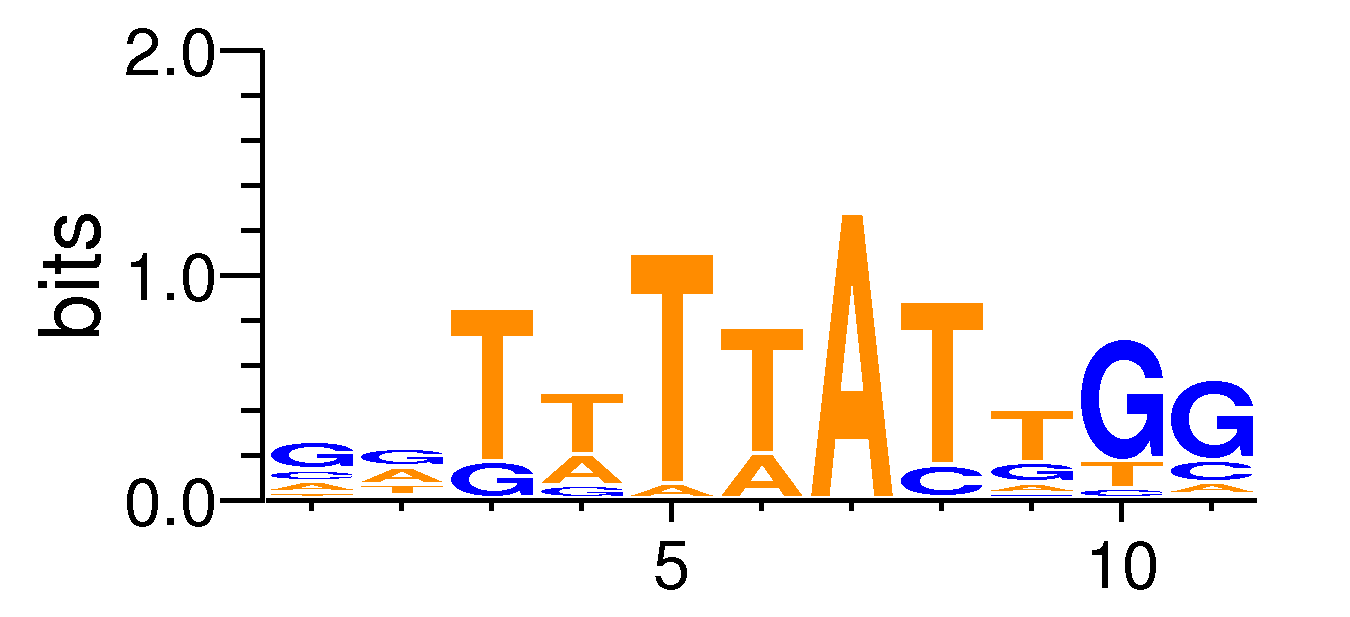
| ExpandMotif matrix |
|---|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| cad#MA0216.1 | -3 | 0.000256908 | 0.122288 | 0.23648 | 7 | GGTTTTATTGG | TTTATTG | + |
| CG4328#MA0182.1 | -3 | 0.000505102 | 0.240429 | 0.23648 | 7 | GGTTTTATTGG | TTTATTA | + |
| HAP5#MA0316.1 | 3 | 0.000936481 | 0.445765 | 0.292013 | 11 | GGTTTTATTGG | TGGGATCTGATTGGT | + |
| Dr#MA0188.1 | -4 | 0.00131132 | 0.624188 | 0.292013 | 7 | GGTTTTATTGG | TAATTGG | - |
| Vsx2#MA0180.1 | -2 | 0.00192777 | 0.917619 | 0.292013 | 9 | GGTTTTATTGG | TTTAATTAG | + |
| CG34031#MA0444.1 | -3 | 0.00262007 | 1.24715 | 0.292013 | 7 | GGTTTTATTGG | TTAATTG | + |
| HAP4#MA0315.1 | 3 | 0.00270974 | 1.28984 | 0.292013 | 11 | GGTTTTATTGG | TCGCCTCTGATTGGT | - |
| B-H2#MA0169.1 | -3 | 0.00280673 | 1.336 | 0.292013 | 7 | GGTTTTATTGG | TTAATTG | + |
| tup#MA0248.1 | -3 | 0.00280673 | 1.336 | 0.292013 | 7 | GGTTTTATTGG | TTAATTG | + |
| CG11085#MA0171.1 | -3 | 0.00389302 | 1.85308 | 0.294195 | 7 | GGTTTTATTGG | TTAATTG | + |
| Abd-B#MA0165.1 | -3 | 0.0041476 | 1.97426 | 0.294195 | 7 | GGTTTTATTGG | TTTATGA | + |
| Hmx#MA0192.1 | -3 | 0.00500076 | 2.38036 | 0.294195 | 7 | GGTTTTATTGG | TTAATTG | + |
| H2.0#MA0448.1 | -3 | 0.00500076 | 2.38036 | 0.294195 | 7 | GGTTTTATTGG | TTAATTA | + |
| CG15696#MA0176.1 | -3 | 0.005318 | 2.53137 | 0.294195 | 7 | GGTTTTATTGG | TTAATTA | + |
| NK7.1#MA0196.1 | -3 | 0.005318 | 2.53137 | 0.294195 | 7 | GGTTTTATTGG | TTAATTG | + |
| Ubx#MA0094.2 | -2 | 0.00564127 | 2.68524 | 0.294195 | 8 | GGTTTTATTGG | TTTAATTA | + |
| Nobox#MA0125.1 | -4 | 0.00564127 | 2.68524 | 0.294195 | 7 | GGTTTTATTGG | TAATTGGT | + |
| hb#MA0049.1 | 0 | 0.00573974 | 2.73212 | 0.294195 | 10 | GGTTTTATTGG | TTTTTTATGC | - |
| CG42234#MA0174.1 | -3 | 0.00600607 | 2.85889 | 0.294195 | 7 | GGTTTTATTGG | TTTATTA | + |
| SMP1#MA0383.1 | 4 | 0.00628377 | 2.99107 | 0.294195 | 11 | GGTTTTATTGG | TGCTAATTTAATTATAGGTAA | - |
| unc-4#MA0250.1 | -3 | 0.00677156 | 3.22326 | 0.301936 | 7 | GGTTTTATTGG | TTAATTG | + |
| YHP1#MA0426.1 | -4 | 0.00745941 | 3.55068 | 0.310107 | 6 | GGTTTTATTGG | TAATTG | + |
| Nkx2-5#MA0063.1 | -3 | 0.00761719 | 3.62578 | 0.310107 | 7 | GGTTTTATTGG | TTAATTG | + |
| CG13424#MA0175.1 | -3 | 0.00854514 | 4.06749 | 0.33339 | 7 | GGTTTTATTGG | TTAATTG | + |
| B-H1#MA0168.1 | -3 | 0.00904181 | 4.3039 | 0.338657 | 7 | GGTTTTATTGG | TTAATTG | + |
| bsh#MA0214.1 | -3 | 0.0101054 | 4.81015 | 0.350455 | 7 | GGTTTTATTGG | TTAATTG | + |
| slou#MA0245.1 | -3 | 0.0101054 | 4.81015 | 0.350455 | 7 | GGTTTTATTGG | TTAATTA | + |
| C15#MA0170.1 | -3 | 0.0125459 | 5.97186 | 0.419555 | 7 | GGTTTTATTGG | TTAATTA | + |
| TBP#MA0108.2 | 5 | 0.0143725 | 6.84131 | 0.454098 | 10 | GGTTTTATTGG | CCCCGCCTTTTATAC | - |
| ARID3A#MA0151.1 | -2 | 0.0148312 | 7.05964 | 0.454098 | 6 | GGTTTTATTGG | TTTAAT | - |
| TBP#MA0108.2 | 5 | 0.0150337 | 7.15605 | 0.454098 | 10 | GGTTTTATTGG | CCCCGCCTTTTATAC | - |
| hbn#MA0226.1 | -3 | 0.0163086 | 7.76291 | 0.476466 | 7 | GGTTTTATTGG | TTAATTA | + |
| HAP3#MA0314.1 | -2 | 0.0171831 | 8.17914 | 0.476466 | 9 | GGTTTTATTGG | TCTGATTGGTTCAGA | + |
| NFYA#MA0060.1 | 0 | 0.0173008 | 8.23518 | 0.476466 | 11 | GGTTTTATTGG | GCGCTGATTGGCTGAG | - |
| CG32532#MA0179.1 | -3 | 0.0180753 | 8.60383 | 0.483572 | 7 | GGTTTTATTGG | TTAATTA | + |
| PHO2#MA0356.1 | -2 | 0.0197944 | 9.42215 | 0.505538 | 6 | GGTTTTATTGG | TATTAT | - |
| ATHB-5#MA0110.1 | -2 | 0.0200151 | 9.52719 | 0.505538 | 9 | GGTTTTATTGG | AATAATTGG | - |
| Oct#MA0197.1 | -2 | 0.020516 | 9.76561 | 0.505538 | 8 | GGTTTTATTGG | TTTAATTA | + |