MacroAPE 1083:Motif516
From FANTOM5_SSTAR
Full Name: motif516
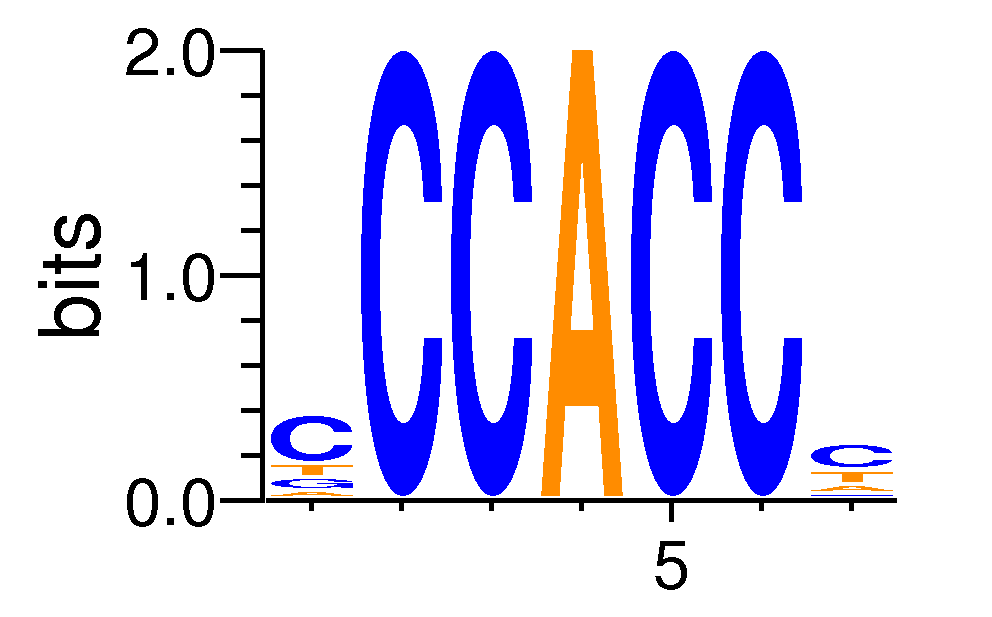
| ExpandMotif matrix |
|---|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| Klf4#MA0039.2 | 2 | 0.00062838 | 0.299109 | 0.502562 | 7 | CCCACCC | GCCCCACCCA | - |
| YPR022C#MA0436.1 | 1 | 0.00106198 | 0.505501 | 0.502562 | 6 | CCCACCC | CCCCACG | + |
| SP1#MA0079.2 | 1 | 0.00266454 | 1.26832 | 0.557287 | 7 | CCCACCC | CCCCGCCCCC | + |
| RPN4#MA0373.1 | 1 | 0.00289904 | 1.37994 | 0.557287 | 6 | CCCACCC | CGCCACC | - |
| AFT2#MA0270.1 | 0 | 0.00294404 | 1.40136 | 0.557287 | 7 | CCCACCC | CACACCCC | + |
| YGR067C#MA0425.1 | 2 | 0.00356781 | 1.69828 | 0.562801 | 7 | CCCACCC | ACCCCACTTTTTTG | + |
| SP1#MA0079.2 | 2 | 0.00449654 | 2.14035 | 0.570003 | 7 | CCCACCC | ACCCCGCCCC | - |
| RREB1#MA0073.1 | 6 | 0.00539764 | 2.56928 | 0.570003 | 7 | CCCACCC | CCCCAAACCACCCCCCCCCC | + |
| YML081W#MA0431.1 | 2 | 0.00625197 | 2.97594 | 0.570003 | 7 | CCCACCC | ACCCCGCAC | + |
| Egr1#MA0162.1 | 3 | 0.00667974 | 3.17956 | 0.570003 | 7 | CCCACCC | CCGCCCACGCA | - |
| UGA3#MA0410.1 | 1 | 0.00675645 | 3.21607 | 0.570003 | 7 | CCCACCC | TCCCGCCG | - |
| MIG2#MA0338.1 | 1 | 0.00722694 | 3.44002 | 0.570003 | 6 | CCCACCC | CCCCGCA | + |
| MIG3#MA0339.1 | 1 | 0.00838427 | 3.99091 | 0.608641 | 6 | CCCACCC | CCCCGCA | + |
| Macho-1#MA0118.1 | 1 | 0.00905291 | 4.30918 | 0.608641 | 7 | CCCACCC | GAACCCCCA | - |
| ACE2#MA0267.1 | 0 | 0.00964601 | 4.5915 | 0.608641 | 7 | CCCACCC | ACCAGCA | + |
| ADR1#MA0268.1 | 2 | 0.0121192 | 5.76872 | 0.716898 | 5 | CCCACCC | ACCCCAC | + |
| btd#MA0443.1 | 0 | 0.013586 | 6.46692 | 0.73526 | 7 | CCCACCC | TCCGCCCCCT | - |
| MET31#MA0333.1 | 1 | 0.0156075 | 7.42918 | 0.73526 | 7 | CCCACCC | CGCCACACT | - |
| opa#MA0456.1 | 1 | 0.0159105 | 7.57338 | 0.73526 | 7 | CCCACCC | GACCCCCCGCTG | + |
| DAL81#MA0290.1 | 3 | 0.0165417 | 7.87384 | 0.73526 | 7 | CCCACCC | AATCCCGCCCGCGGCTTTT | - |
| ZNF354C#MA0130.1 | 1 | 0.0172677 | 8.21943 | 0.73526 | 5 | CCCACCC | ATCCAC | + |
| Lag1#MA0193.1 | -1 | 0.0172996 | 8.2346 | 0.73526 | 6 | CCCACCC | CTACCAA | + |
| Pax4#MA0068.1 | 22 | 0.0185535 | 8.83145 | 0.73526 | 7 | CCCACCC | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| Su(H)#MA0085.1 | 9 | 0.0190849 | 9.08441 | 0.73526 | 7 | CCCACCC | ATCTCGGTTCCCACAA | - |
| Nkx3-2#MA0122.1 | 0 | 0.0194212 | 9.24451 | 0.73526 | 7 | CCCACCC | TCCACTTAA | - |