MacroAPE 1083:PAX5 motifB:GM12878-PAX5-ChIP-Seq/Homer
From FANTOM5_SSTAR
Full Name: PAX5_motifB:GM12878-PAX5-ChIP-Seq/Homer
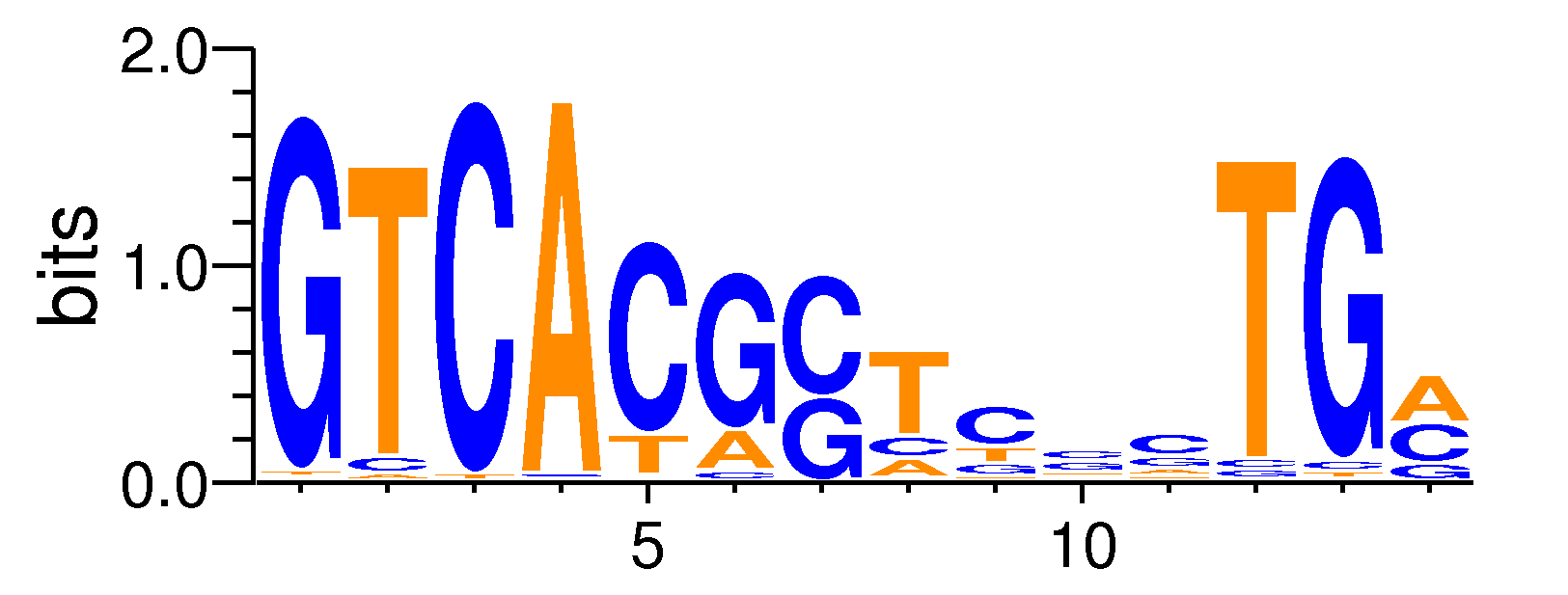
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| Pax2#MA0067.1 | 1 | 0.000145698 | 0.0693522 | 0.138669 | 7 | GTCACGCTCCCTGA | AGTCACGG | + |
| usp#MA0016.1 | 3 | 0.00045733 | 0.217689 | 0.217634 | 7 | GTCACGCTCCCTGA | GGGGTCACGG | + |
| hkb#MA0450.1 | -1 | 0.00154768 | 0.736694 | 0.491005 | 9 | GTCACGCTCCCTGA | TCACGCCCC | - |
| NFE2L1::MafG#MA0089.1 | 0 | 0.00345219 | 1.64324 | 0.734322 | 6 | GTCACGCTCCCTGA | GTCATG | - |
| Pax5#MA0014.1 | 2 | 0.00385771 | 1.83627 | 0.734322 | 14 | GTCACGCTCCCTGA | CCGCTACGCTTCAGTGCTCT | - |
| ABF1#MA0265.1 | 2 | 0.00590429 | 2.81044 | 0.799398 | 14 | GTCACGCTCCCTGA | ATATCACTTTATACGA | - |
| ESR2#MA0258.1 | 4 | 0.00801178 | 3.81361 | 0.799398 | 14 | GTCACGCTCCCTGA | CAAGGTCACGGTGACCTG | + |
| PPARG#MA0066.1 | 4 | 0.0095093 | 4.52643 | 0.799398 | 14 | GTCACGCTCCCTGA | GTAGGTCACGGTGACCTACT | + |
| ESR1#MA0112.1 | 3 | 0.0103693 | 4.93576 | 0.799398 | 14 | GTCACGCTCCCTGA | GGGGTCACGGTGACCTGG | - |
| MET31#MA0333.1 | 1 | 0.0110697 | 5.26919 | 0.799398 | 8 | GTCACGCTCCCTGA | CGCCACACT | - |
| ARG81#MA0272.1 | 3 | 0.0120537 | 5.73757 | 0.799398 | 5 | GTCACGCTCCCTGA | AGAGTCAC | - |
| CREB1#MA0018.1 | 3 | 0.0124223 | 5.91301 | 0.799398 | 9 | GTCACGCTCCCTGA | GACGTCACCAGG | - |
| h#MA0449.1 | 0 | 0.0126281 | 6.01099 | 0.799398 | 10 | GTCACGCTCCCTGA | GGCACGTGCC | + |
| NR4A2#MA0160.1 | 3 | 0.0127076 | 6.0488 | 0.799398 | 5 | GTCACGCTCCCTGA | AAGGTCAC | + |
| Arnt::Ahr#MA0006.1 | -2 | 0.0136402 | 6.49271 | 0.799398 | 6 | GTCACGCTCCCTGA | CACGCA | - |
| GCN4#MA0303.1 | 10 | 0.0142786 | 6.7966 | 0.799398 | 11 | GTCACGCTCCCTGA | CAAGGGATGAGTCATACTTCA | + |