MacroAPE 1083:SOX18 f1
From FANTOM5_SSTAR
Full Name: SOX18_f1
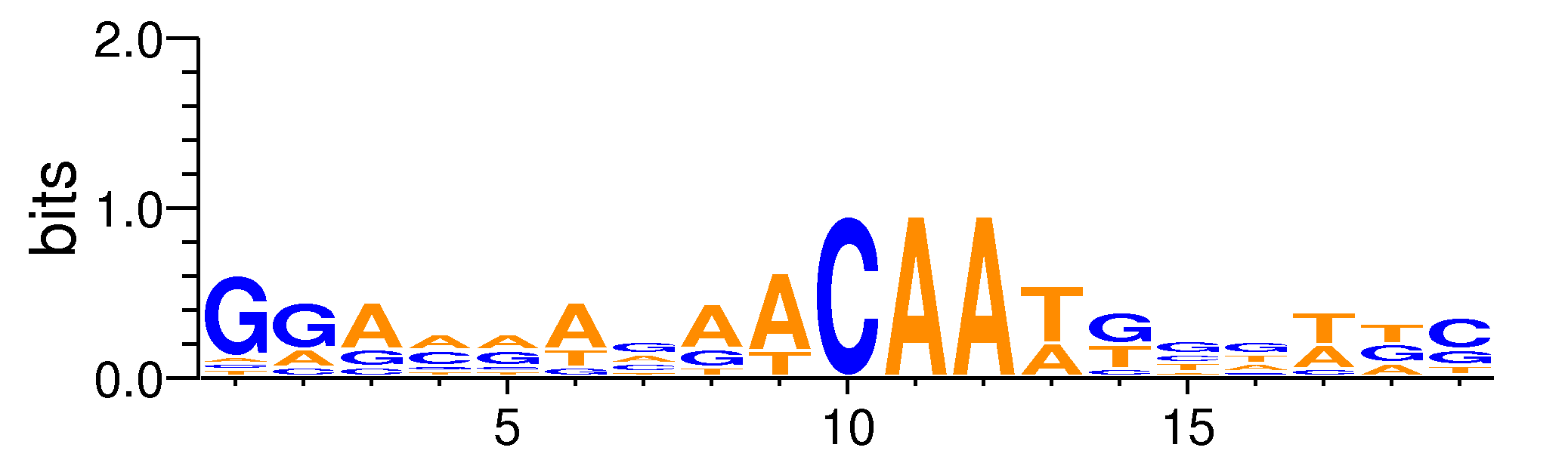
| ExpandMotif matrix |
|---|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| D#MA00445.1 | -5 | 7.66241e-06 | 0.00364731 | 0.00594802 | 11 | GGAAAAGAACAATGGGTTC | AGAACAATGGA | - |
| SOX9#MA0077.1 | -6 | 1.26951e-05 | 0.00604288 | 0.00594802 | 9 | GGAAAAGAACAATGGGTTC | GAACAATGG | + |
| SRY#MA0084.1 | -4 | 3.42782e-05 | 0.0163164 | 0.00921693 | 9 | GGAAAAGAACAATGGGTTC | GTAAACAAT | + |
| ROX1#MA0371.1 | -4 | 3.93442e-05 | 0.0187279 | 0.00921693 | 12 | GGAAAAGAACAATGGGTTC | GAGAACAATGGA | - |
| Sox5#MA0087.1 | -6 | 0.000159547 | 0.0759444 | 0.0299009 | 7 | GGAAAAGAACAATGGGTTC | AAACAAT | + |
| Sox17#MA0078.1 | -7 | 0.00050187 | 0.23889 | 0.07838 | 9 | GGAAAAGAACAATGGGTTC | GACAATGAG | - |
| HCM1#MA0317.1 | -4 | 0.000912696 | 0.434443 | 0.122178 | 8 | GGAAAAGAACAATGGGTTC | ATAAACAA | + |
| id1#MA0120.1 | 0 | 0.00139441 | 0.663739 | 0.148446 | 12 | GGAAAAGAACAATGGGTTC | CGAAAAGGAAAA | - |
| SOX10#MA0442.1 | -8 | 0.00142576 | 0.678663 | 0.148446 | 6 | GGAAAAGAACAATGGGTTC | ACAAAG | - |
| br_Z1#MA0010.1 | -1 | 0.00237772 | 1.13179 | 0.214324 | 14 | GGAAAAGAACAATGGGTTC | GTAATAAACAAATC | + |
| FOXF2#MA0030.1 | 1 | 0.00251592 | 1.19758 | 0.214324 | 13 | GGAAAAGAACAATGGGTTC | CAAACGTAAACAAT | + |
| Foxd3#MA0041.1 | -2 | 0.00307459 | 1.46351 | 0.238583 | 12 | GGAAAAGAACAATGGGTTC | AAACAAACATTC | - |
| STB3#MA0390.1 | 1 | 0.00330991 | 1.57552 | 0.238583 | 19 | GGAAAAGAACAATGGGTTC | CCAGAGTGAAAAATTTTGGAC | - |
| slp1#MA0458.1 | -1 | 0.00381958 | 1.81812 | 0.255654 | 11 | GGAAAAGAACAATGGGTTC | AATGTAAACAA | - |
| MGA1#MA0336.1 | 3 | 0.00520795 | 2.47898 | 0.325343 | 18 | GGAAAAGAACAATGGGTTC | AGGACTATAGAACACTCTAAA | + |
| SFL1#MA0377.1 | 3 | 0.0057476 | 2.73586 | 0.329311 | 18 | GGAAAAGAACAATGGGTTC | TAGAGAATAGAAGAAATAAAA | + |
| FKH1#MA0296.1 | 2 | 0.00597433 | 2.84378 | 0.329311 | 18 | GGAAAAGAACAATGGGTTC | TGAATTGTAAACAAAGAGGA | + |
| Pax4#MA0068.1 | 6 | 0.00685056 | 3.26087 | 0.356631 | 19 | GGAAAAGAACAATGGGTTC | GGGGGGGGAGTGGAGTATTGGAAATTTTTC | - |
| SFP1#MA0378.1 | -2 | 0.00806774 | 3.84024 | 0.397891 | 17 | GGAAAAGAACAATGGGTTC | CCGTAGAAAATTTTTTTCAAT | - |
| pan#MA0237.1 | -6 | 0.00867995 | 4.13166 | 0.40668 | 8 | GGAAAAGAACAATGGGTTC | GATCAAAG | - |
| Sox2#MA0143.1 | 0 | 0.00954967 | 4.54565 | 0.406754 | 15 | GGAAAAGAACAATGGGTTC | TTTGCATAACAAAGG | - |
| En1#MA0027.1 | -4 | 0.0112053 | 5.33371 | 0.442521 | 11 | GGAAAAGAACAATGGGTTC | GAACACTACTT | - |
| Ar#MA0007.1 | -2 | 0.0121909 | 5.80287 | 0.442521 | 17 | GGAAAAGAACAATGGGTTC | ATAAGAACACCCTGTACCCGCC | + |
| Foxa2#MA0047.1 | -2 | 0.0122784 | 5.84452 | 0.442521 | 12 | GGAAAAGAACAATGGGTTC | AAGTAAATATTG | - |
| Foxq1#MA0040.1 | -3 | 0.0140057 | 6.6667 | 0.464685 | 11 | GGAAAAGAACAATGGGTTC | AATAAACAATA | - |
| SPT23#MA0388.1 | -4 | 0.0142139 | 6.76579 | 0.464685 | 8 | GGAAAAGAACAATGGGTTC | GAAATCAA | + |
| br_Z3#MA0012.1 | -5 | 0.0147962 | 7.04297 | 0.464685 | 11 | GGAAAAGAACAATGGGTTC | TAAACTAAAAG | + |
| NFYA#MA0060.1 | -3 | 0.0150679 | 7.17231 | 0.464685 | 16 | GGAAAAGAACAATGGGTTC | CTCAGCCAATCAGCGC | + |
| NRG1#MA0347.1 | 0 | 0.0153729 | 7.31748 | 0.464685 | 19 | GGAAAAGAACAATGGGTTC | TGCGATGGACCCTGATCTAG | - |
| NR3C1#MA0113.1 | -4 | 0.017902 | 8.52137 | 0.511269 | 15 | GGAAAAGAACAATGGGTTC | GGGAACATTATGTCCTAT | + |
| MATA1#MA0327.1 | -6 | 0.0180052 | 8.57047 | 0.511269 | 7 | GGAAAAGAACAATGGGTTC | ACACAAT | + |
| HMG-1#MA0044.1 | -4 | 0.0187504 | 8.92521 | 0.516771 | 9 | GGAAAAGAACAATGGGTTC | GAATACAAC | - |